
Mir Lab
@mirlab.bsky.social
Lab run by Mustafa Mir studying transcription and nuclear organization using live imaging at University of Pennsylvania, Children's Hospital of Philadelphia, and Howard Hughes Medical Institute. mir-lab.com
Reposted by Mir Lab
Huge props to our grad student @manyakapoor.bsky.social for developing the simulations and the new MS2 analysis amongst other things! We also thank the transcription community for their invaluable feedback and always welcome further discussion😊
September 18, 2025 at 3:24 PM
Huge props to our grad student @manyakapoor.bsky.social for developing the simulations and the new MS2 analysis amongst other things! We also thank the transcription community for their invaluable feedback and always welcome further discussion😊
5/n: Please reach out directly to me if you are interested. This is a unique opportunity at a time where increased centralized resources are critical to maintain and accelerate the pace of scientific discovery.
April 11, 2025 at 10:48 AM
5/n: Please reach out directly to me if you are interested. This is a unique opportunity at a time where increased centralized resources are critical to maintain and accelerate the pace of scientific discovery.
4/n We are seeking a microscope specialist/engineer. The role will include developing and implementing new microscope hardware, control software, customized analysis pipelines as well as collaborating with core users.
April 11, 2025 at 10:48 AM
4/n We are seeking a microscope specialist/engineer. The role will include developing and implementing new microscope hardware, control software, customized analysis pipelines as well as collaborating with core users.
3/n We will provide custom built ad hoc optical microscopy solutions, advanced image analysis, and consultation services to researchers across campus and beyond. We will implement technology in response to experimental needs instead of working within the bounds of commercially available technology.
Advanced Core for Microscope Engineering (ACME) | Advanced Core for Microscope Engineering | Perelman School of Medicine at the University of Pennsylvania
www.med.upenn.edu
April 11, 2025 at 10:48 AM
3/n We will provide custom built ad hoc optical microscopy solutions, advanced image analysis, and consultation services to researchers across campus and beyond. We will implement technology in response to experimental needs instead of working within the bounds of commercially available technology.
2/n: ACME is co-directed by myself (Mustafa Mir), Melike Lakadamyali @melikel.bsky.social (www.lakadamyali-lab.com), Yihui Shen (www.imaging-systems-metabolism-lab.com), and Andrea Stout @aleestoutphd.bsky.social (www.med.upenn.edu/cdbmicroscopycore)
April 11, 2025 at 10:48 AM
2/n: ACME is co-directed by myself (Mustafa Mir), Melike Lakadamyali @melikel.bsky.social (www.lakadamyali-lab.com), Yihui Shen (www.imaging-systems-metabolism-lab.com), and Andrea Stout @aleestoutphd.bsky.social (www.med.upenn.edu/cdbmicroscopycore)
9/ We welcome feedback, questions, and critical discussion from the community. Huge thanks to all our amazing co-authors for their insight, support, and collaboration throughout this work.
April 9, 2025 at 2:31 PM
9/ We welcome feedback, questions, and critical discussion from the community. Huge thanks to all our amazing co-authors for their insight, support, and collaboration throughout this work.
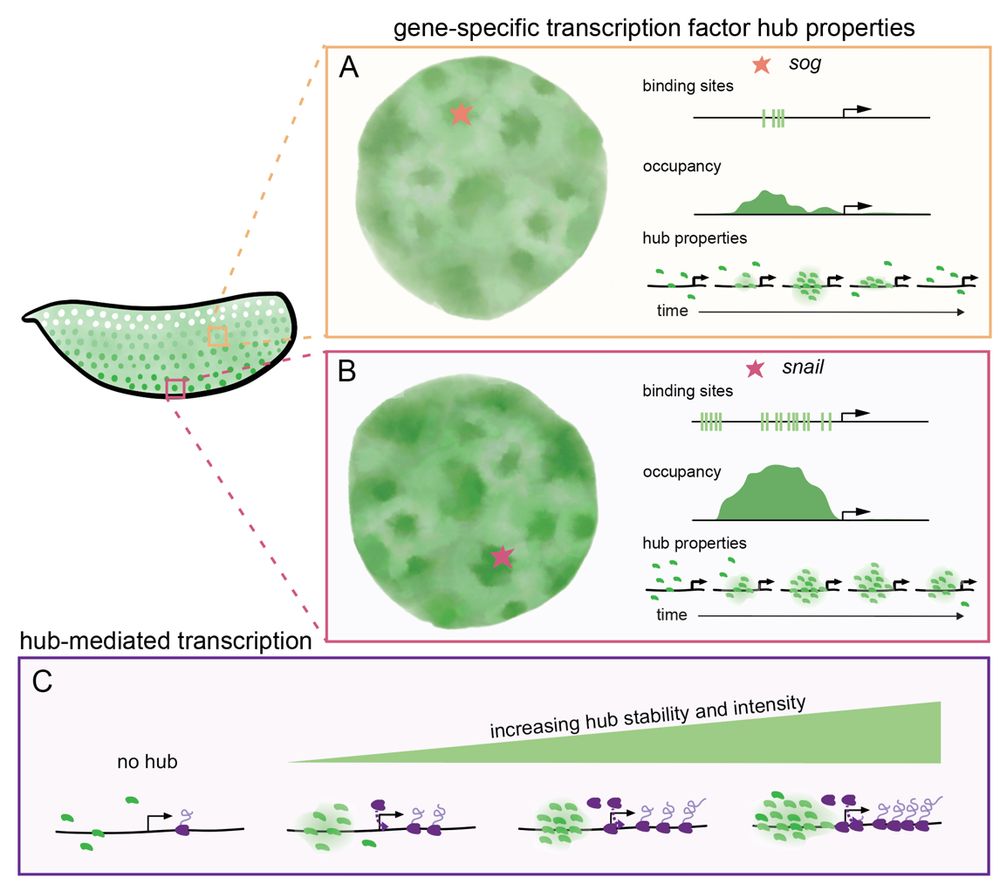
8/n: We suggest that enhancers encode the biophysical properties of transcription factor hubs (intensity and stability) through binding affinities and cofactors. Hub properties dictate gene occupancy and transcriptional kinetics, suggesting hubs are active executors of enhancer-encoded regulation.

April 9, 2025 at 2:31 PM
8/n: We suggest that enhancers encode the biophysical properties of transcription factor hubs (intensity and stability) through binding affinities and cofactors. Hub properties dictate gene occupancy and transcriptional kinetics, suggesting hubs are active executors of enhancer-encoded regulation.
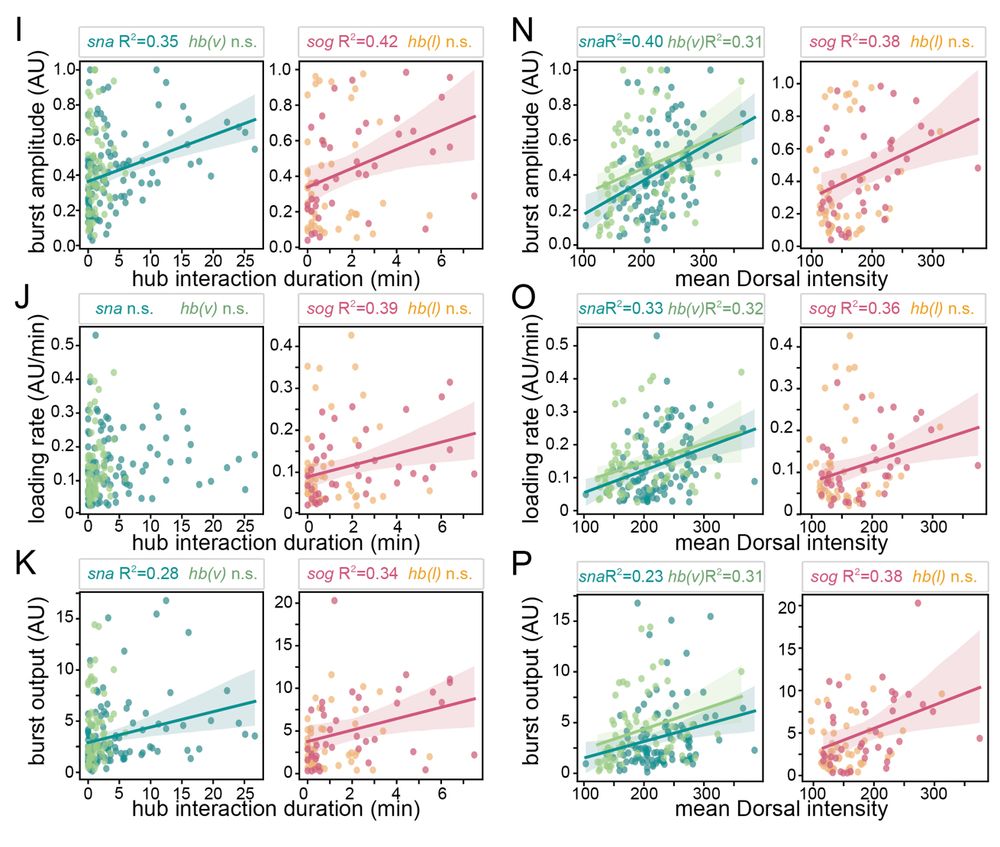
7/n: Hub intensity and duration correlate with burst amplitude, loading rate, and output suggesting that persistently interacting hubs with more molecules tune gene expression. Consistent with the idea that hubs facilitate transcription factor binding and promote expression.
April 9, 2025 at 2:31 PM
7/n: Hub intensity and duration correlate with burst amplitude, loading rate, and output suggesting that persistently interacting hubs with more molecules tune gene expression. Consistent with the idea that hubs facilitate transcription factor binding and promote expression.
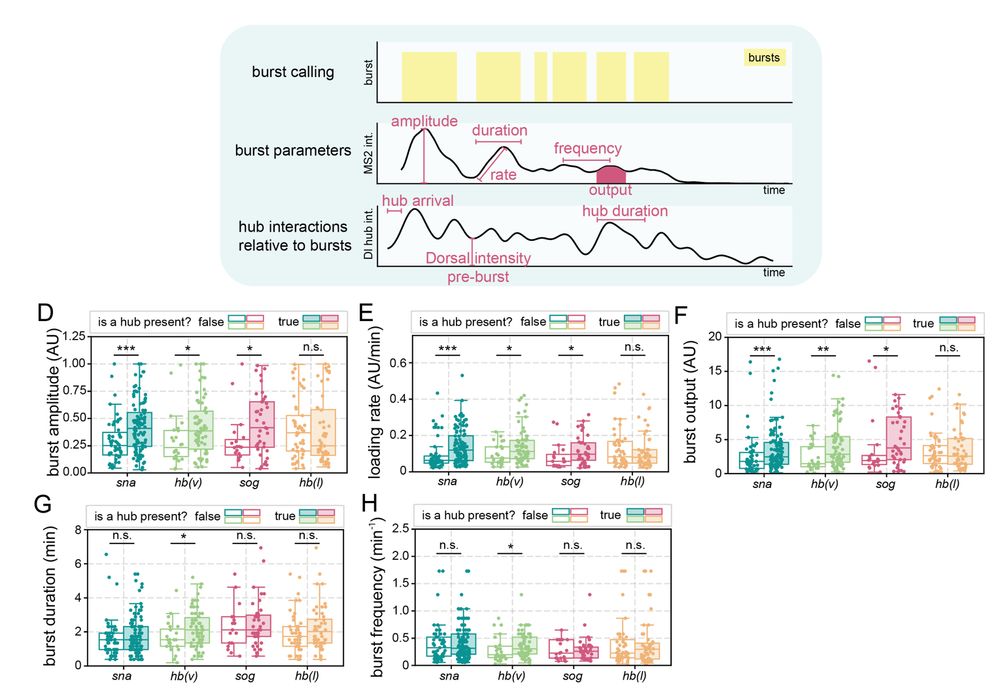
6/n: Hub presence at a gene is correlated with increased transcription burst amplitude, Pol II loading rate, and total output. Surprisingly, transient interactions between Dorsal hubs and hunchback (lacking Dorsal binding) boosts transcription in ventral nuclei where Dorsal concentration is highest
April 9, 2025 at 2:31 PM
6/n: Hub presence at a gene is correlated with increased transcription burst amplitude, Pol II loading rate, and total output. Surprisingly, transient interactions between Dorsal hubs and hunchback (lacking Dorsal binding) boosts transcription in ventral nuclei where Dorsal concentration is highest
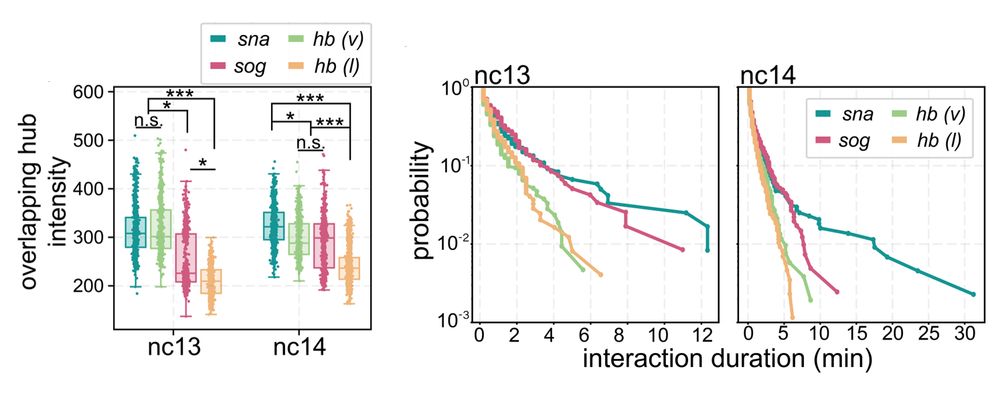
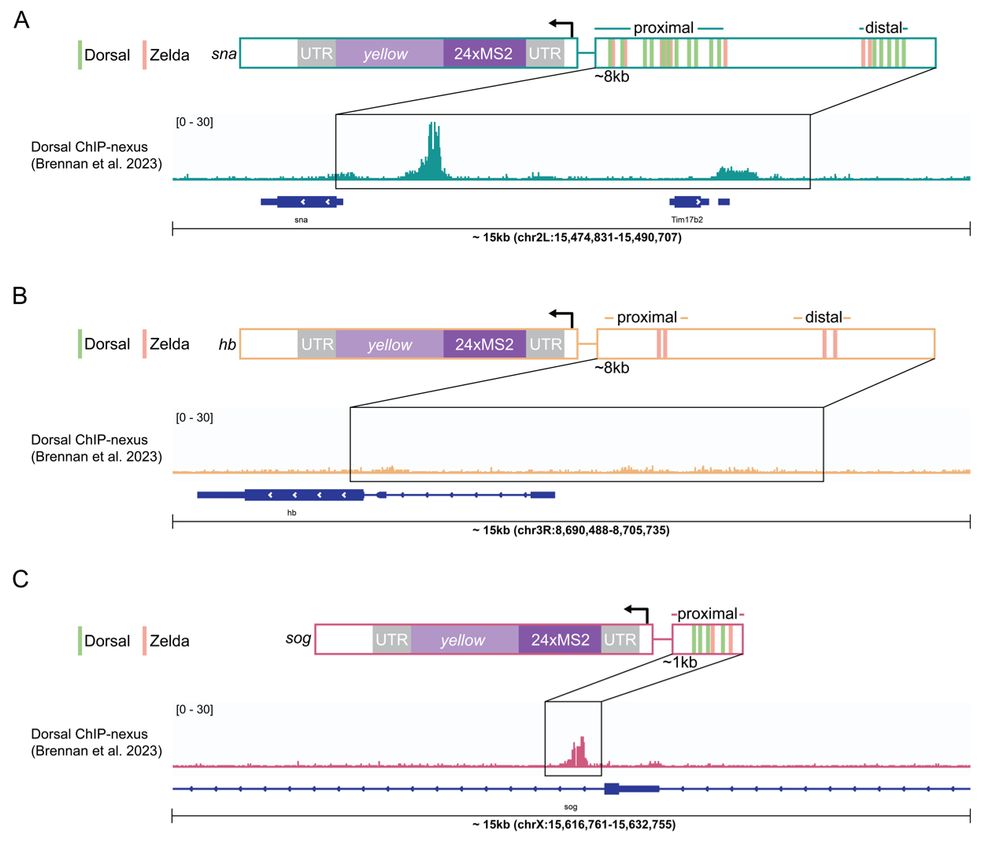
5/n: We found:
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
April 9, 2025 at 2:31 PM
5/n: We found:
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
4/n: Next we examined Dorsal hub interactions at two target genes with distinct Dorsal occupancy and enhancer grammars, snai and sog, and as a negative control examined hunchback which has no dorsal sites.
April 9, 2025 at 2:31 PM
4/n: Next we examined Dorsal hub interactions at two target genes with distinct Dorsal occupancy and enhancer grammars, snai and sog, and as a negative control examined hunchback which has no dorsal sites.
3/n: First, we find that hubs form rapidly after mitosis and last through interphase. Nuclear hub density doesn't depend on nuclear concentration but hub intensity does, suggesting that hub formation may be independent of concentration, but hubs accumulate more protein with increased concentrations.
April 9, 2025 at 2:31 PM
3/n: First, we find that hubs form rapidly after mitosis and last through interphase. Nuclear hub density doesn't depend on nuclear concentration but hub intensity does, suggesting that hub formation may be independent of concentration, but hubs accumulate more protein with increased concentrations.
2/n: Transcription factor hubs (dense local accumulations) regulate gene expression by promoting the frequency of binding. They are often discussed as having uniform properties based on protein sequence and concentration. We wondered if their biophysical properties vary in a gene dependent manner.
April 9, 2025 at 2:31 PM
2/n: Transcription factor hubs (dense local accumulations) regulate gene expression by promoting the frequency of binding. They are often discussed as having uniform properties based on protein sequence and concentration. We wondered if their biophysical properties vary in a gene dependent manner.
And all the MS2 interaction analysis was done by an amazing bioengineering graduate student @manyakapoor.bsky.social who just joined Bsky!
February 12, 2025 at 4:49 PM
And all the MS2 interaction analysis was done by an amazing bioengineering graduate student @manyakapoor.bsky.social who just joined Bsky!
Major oversight in my earlier postdoc: This work was led by our wonderful postdoc @mukherja.bsky.social who also built the microscope that enabled these experiments
February 12, 2025 at 4:18 PM
Major oversight in my earlier postdoc: This work was led by our wonderful postdoc @mukherja.bsky.social who also built the microscope that enabled these experiments
We can't follow the same cluster but see a shift in cluster kinetics across nuclei. We can't say anything about initiation factors since we are just looking at pol2 and we are testing effects of recruitment vs elongation with the drugs. Will check our language to make sure this is clear. Thanks.
February 12, 2025 at 1:56 PM
We can't follow the same cluster but see a shift in cluster kinetics across nuclei. We can't say anything about initiation factors since we are just looking at pol2 and we are testing effects of recruitment vs elongation with the drugs. Will check our language to make sure this is clear. Thanks.
7/n Overall we show that Pol II clusters are functionally labile and suggest that they likely represent collections of molecules engaged at a single gene locus. Please take a read and send us your critical feedback!
February 12, 2025 at 11:36 AM
7/n Overall we show that Pol II clusters are functionally labile and suggest that they likely represent collections of molecules engaged at a single gene locus. Please take a read and send us your critical feedback!
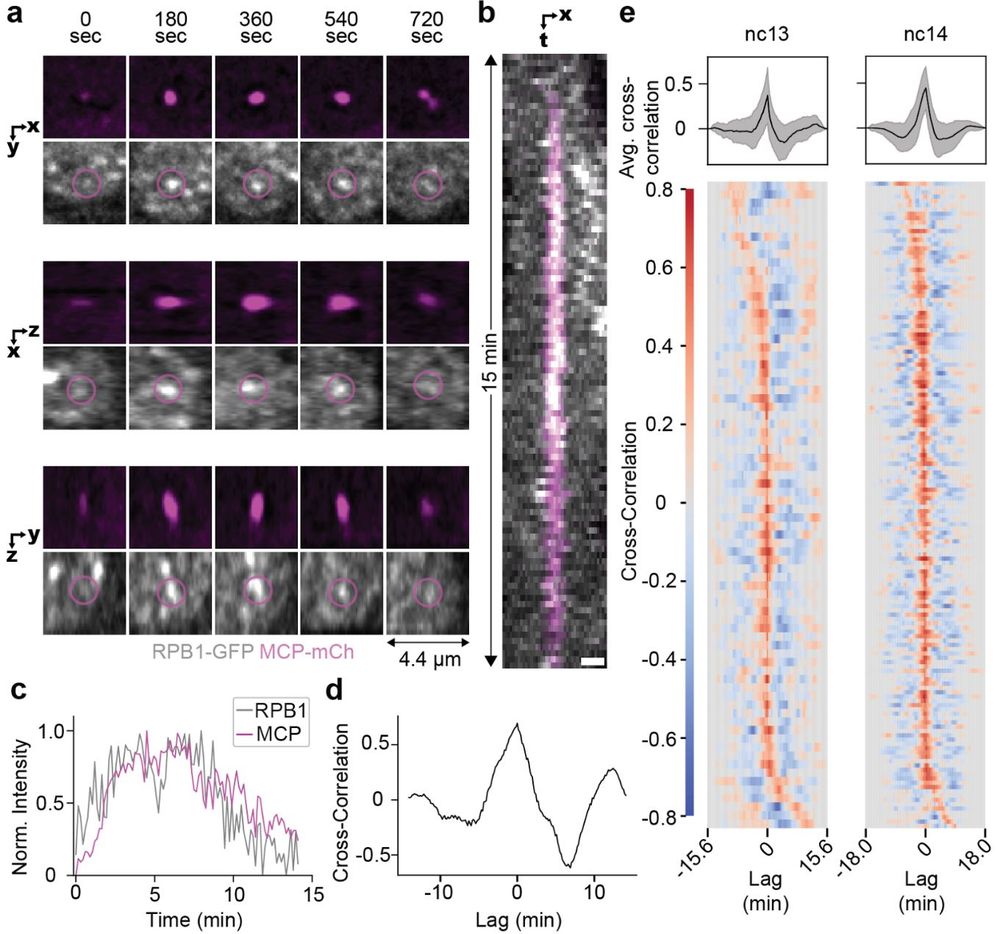
6/n Using simultaneous imaging of nascent transcription and Pol II clusters we found that a single cluster remains associated with an active gene during a transcription burst and the intensity of the cluster is highly correlated in real time with the amount of nascent transcription.
February 12, 2025 at 11:36 AM
6/n Using simultaneous imaging of nascent transcription and Pol II clusters we found that a single cluster remains associated with an active gene during a transcription burst and the intensity of the cluster is highly correlated in real time with the amount of nascent transcription.
5/n Using inhibitors of initiation and elongation we found that cluster formation is dependent on initiation and that transcription elongation leads to a reduction in cluster lifetime.
February 12, 2025 at 11:36 AM
5/n Using inhibitors of initiation and elongation we found that cluster formation is dependent on initiation and that transcription elongation leads to a reduction in cluster lifetime.
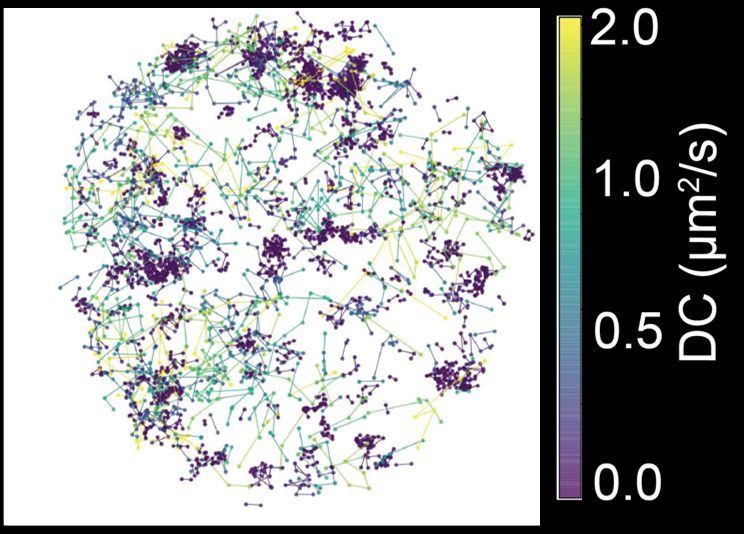
4/n We found that Pol II clusters undergo a transformation as the zygotic genome is activated. From initially being mostly composed of molecules engaged in transcription initiation to becoming hotspots of elongating molecules.
February 12, 2025 at 11:36 AM
4/n We found that Pol II clusters undergo a transformation as the zygotic genome is activated. From initially being mostly composed of molecules engaged in transcription initiation to becoming hotspots of elongating molecules.

