
Mir Lab
@mirlab.bsky.social
Lab run by Mustafa Mir studying transcription and nuclear organization using live imaging at University of Pennsylvania, Children's Hospital of Philadelphia, and Howard Hughes Medical Institute. mir-lab.com
1/n We have an exciting new initiative at UPenn to provide custom advanced microscopy solutions: Advanced Core for Microscope Engineering (ACME): www.med.upenn.edu/cdbacme/ We are looking for a specialist to help run this new facility, job description is attached please help share widely!

April 11, 2025 at 10:48 AM
1/n We have an exciting new initiative at UPenn to provide custom advanced microscopy solutions: Advanced Core for Microscope Engineering (ACME): www.med.upenn.edu/cdbacme/ We are looking for a specialist to help run this new facility, job description is attached please help share widely!
8/n: We suggest that enhancers encode the biophysical properties of transcription factor hubs (intensity and stability) through binding affinities and cofactors. Hub properties dictate gene occupancy and transcriptional kinetics, suggesting hubs are active executors of enhancer-encoded regulation.

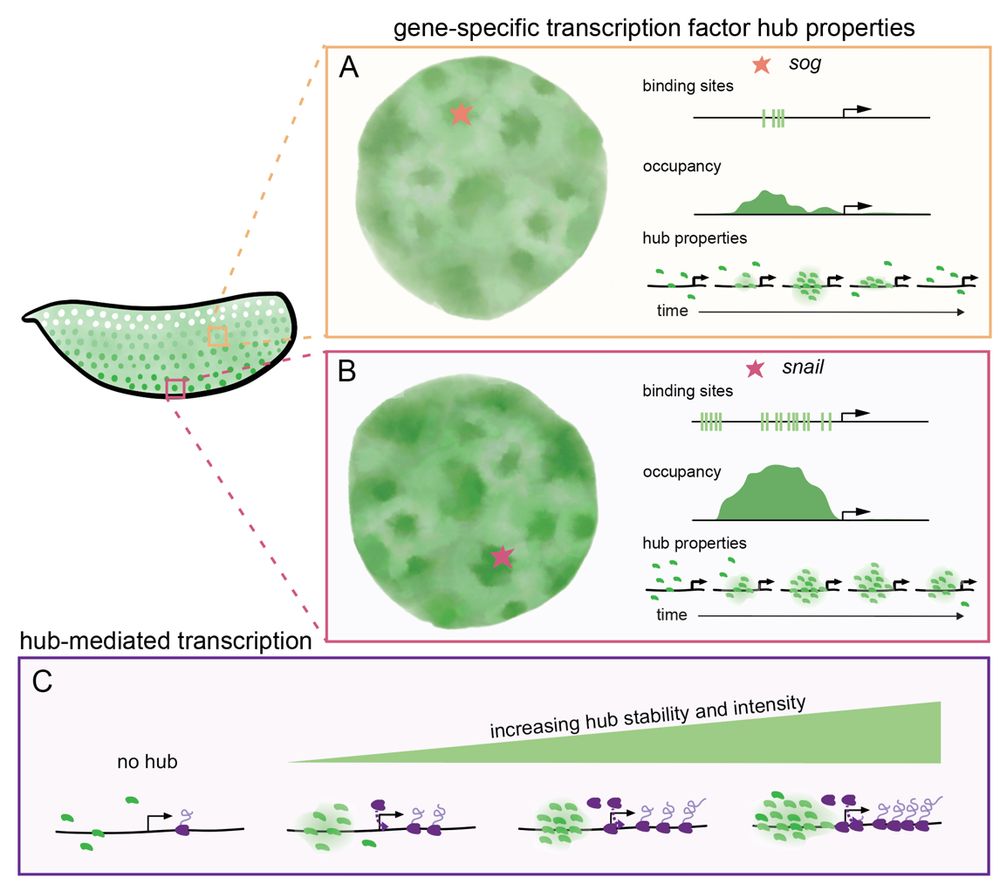
April 9, 2025 at 2:31 PM
8/n: We suggest that enhancers encode the biophysical properties of transcription factor hubs (intensity and stability) through binding affinities and cofactors. Hub properties dictate gene occupancy and transcriptional kinetics, suggesting hubs are active executors of enhancer-encoded regulation.
7/n: Hub intensity and duration correlate with burst amplitude, loading rate, and output suggesting that persistently interacting hubs with more molecules tune gene expression. Consistent with the idea that hubs facilitate transcription factor binding and promote expression.
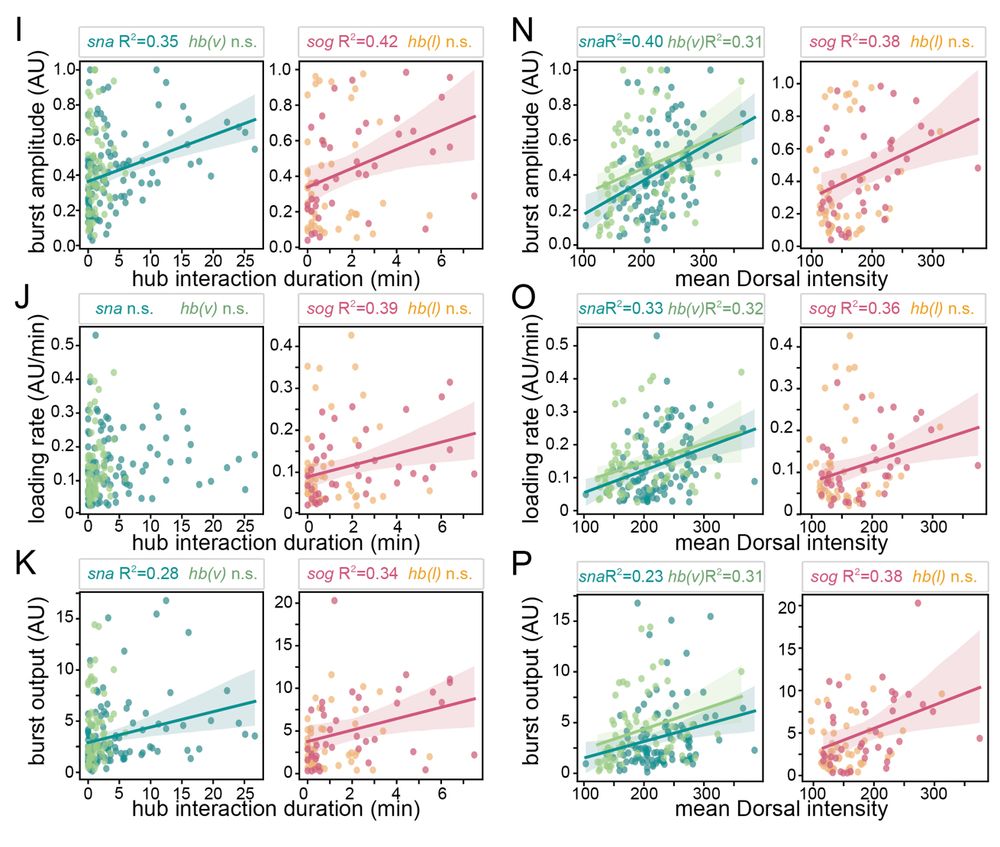
April 9, 2025 at 2:31 PM
7/n: Hub intensity and duration correlate with burst amplitude, loading rate, and output suggesting that persistently interacting hubs with more molecules tune gene expression. Consistent with the idea that hubs facilitate transcription factor binding and promote expression.
6/n: Hub presence at a gene is correlated with increased transcription burst amplitude, Pol II loading rate, and total output. Surprisingly, transient interactions between Dorsal hubs and hunchback (lacking Dorsal binding) boosts transcription in ventral nuclei where Dorsal concentration is highest
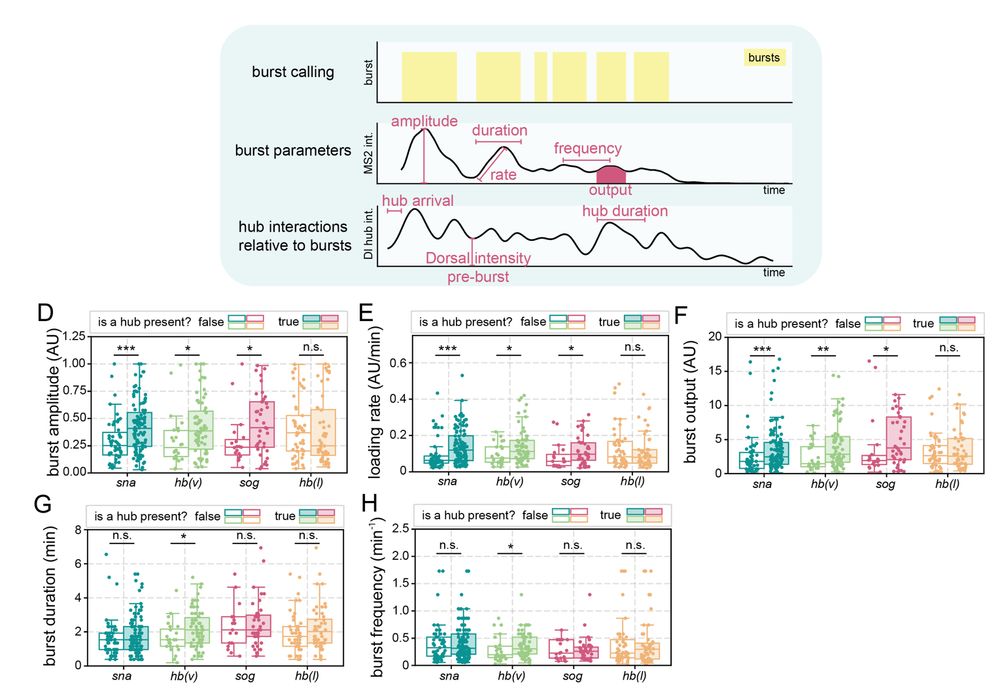
April 9, 2025 at 2:31 PM
6/n: Hub presence at a gene is correlated with increased transcription burst amplitude, Pol II loading rate, and total output. Surprisingly, transient interactions between Dorsal hubs and hunchback (lacking Dorsal binding) boosts transcription in ventral nuclei where Dorsal concentration is highest
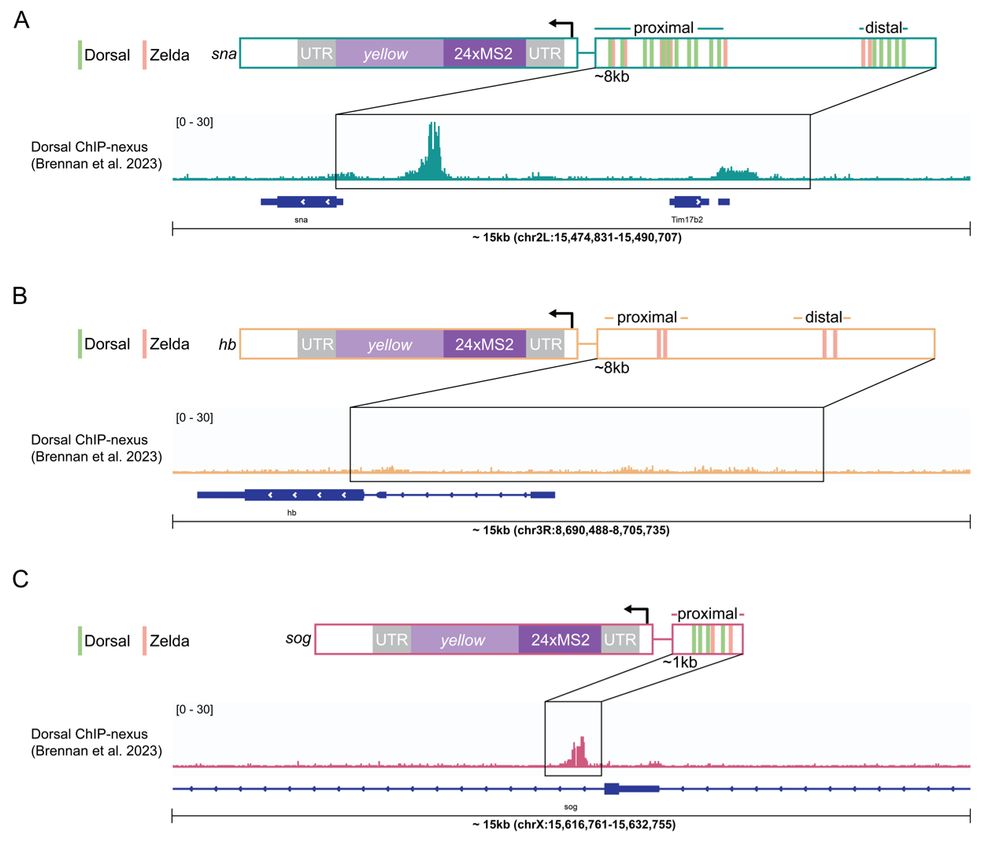
5/n: We found:
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
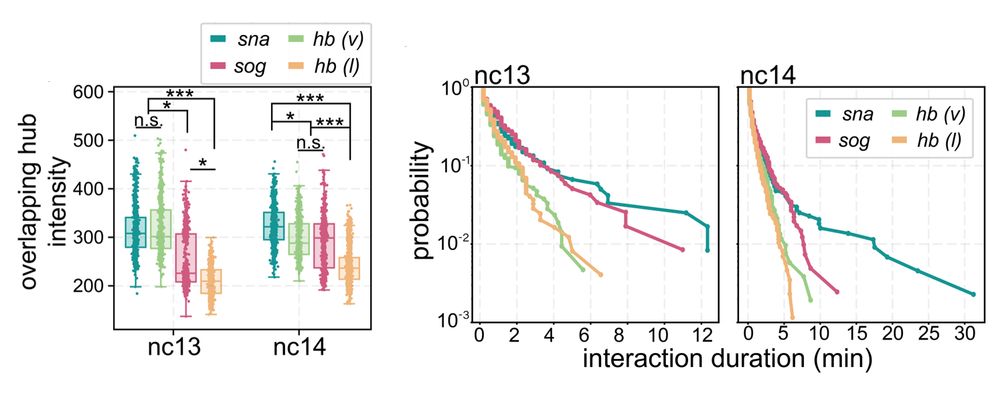
April 9, 2025 at 2:31 PM
5/n: We found:
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
4/n: Next we examined Dorsal hub interactions at two target genes with distinct Dorsal occupancy and enhancer grammars, snai and sog, and as a negative control examined hunchback which has no dorsal sites.
April 9, 2025 at 2:31 PM
4/n: Next we examined Dorsal hub interactions at two target genes with distinct Dorsal occupancy and enhancer grammars, snai and sog, and as a negative control examined hunchback which has no dorsal sites.
3/n: First, we find that hubs form rapidly after mitosis and last through interphase. Nuclear hub density doesn't depend on nuclear concentration but hub intensity does, suggesting that hub formation may be independent of concentration, but hubs accumulate more protein with increased concentrations.
April 9, 2025 at 2:31 PM
3/n: First, we find that hubs form rapidly after mitosis and last through interphase. Nuclear hub density doesn't depend on nuclear concentration but hub intensity does, suggesting that hub formation may be independent of concentration, but hubs accumulate more protein with increased concentrations.
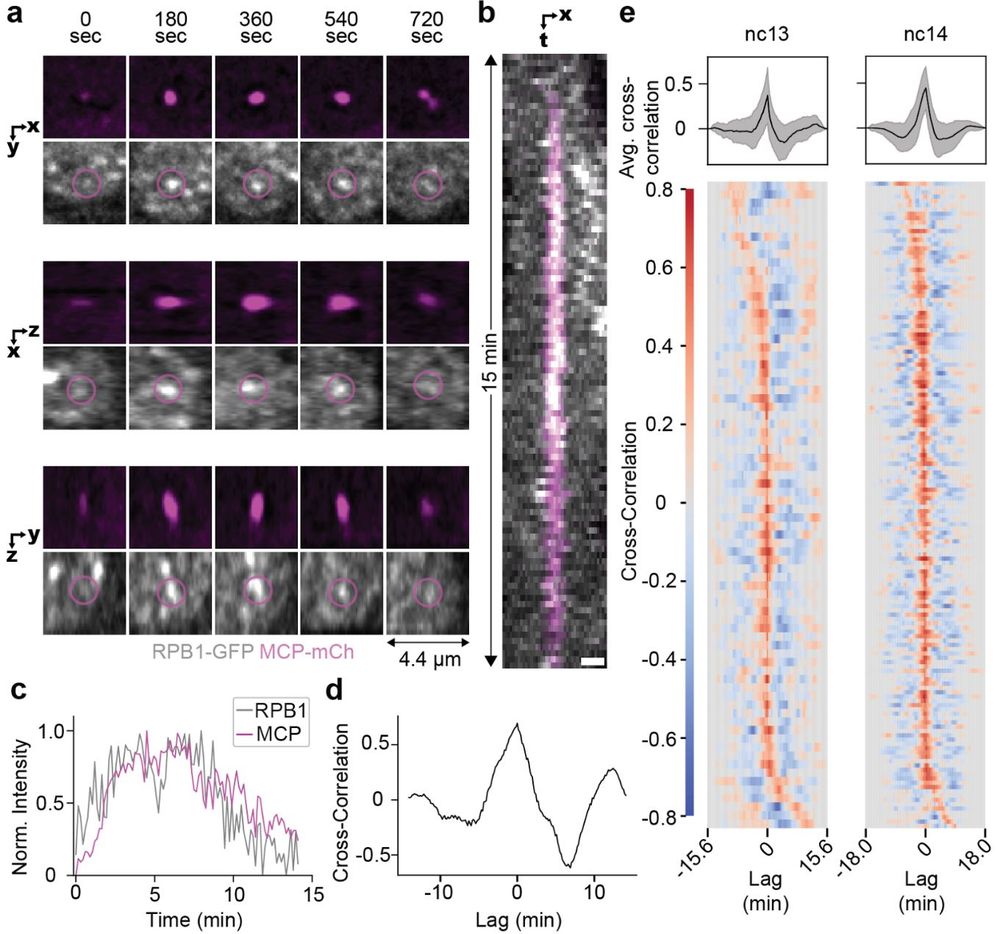
6/n Using simultaneous imaging of nascent transcription and Pol II clusters we found that a single cluster remains associated with an active gene during a transcription burst and the intensity of the cluster is highly correlated in real time with the amount of nascent transcription.
February 12, 2025 at 11:36 AM
6/n Using simultaneous imaging of nascent transcription and Pol II clusters we found that a single cluster remains associated with an active gene during a transcription burst and the intensity of the cluster is highly correlated in real time with the amount of nascent transcription.
5/n Using inhibitors of initiation and elongation we found that cluster formation is dependent on initiation and that transcription elongation leads to a reduction in cluster lifetime.
February 12, 2025 at 11:36 AM
5/n Using inhibitors of initiation and elongation we found that cluster formation is dependent on initiation and that transcription elongation leads to a reduction in cluster lifetime.
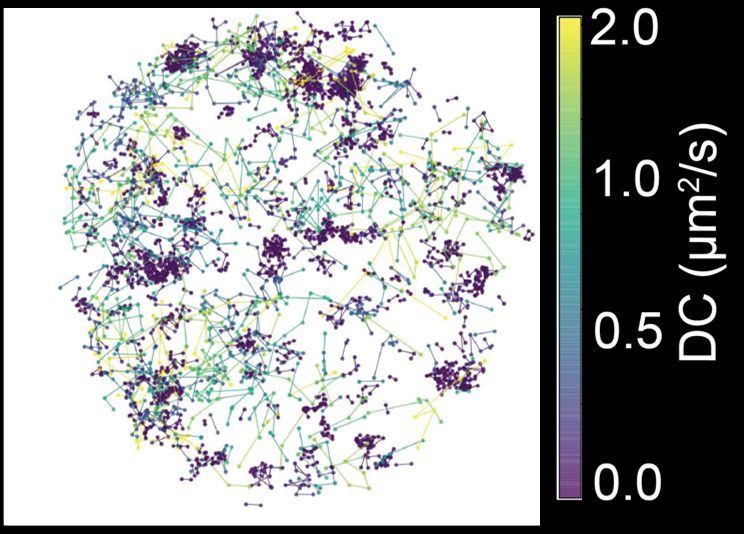
4/n We found that Pol II clusters undergo a transformation as the zygotic genome is activated. From initially being mostly composed of molecules engaged in transcription initiation to becoming hotspots of elongating molecules.
February 12, 2025 at 11:36 AM
4/n We found that Pol II clusters undergo a transformation as the zygotic genome is activated. From initially being mostly composed of molecules engaged in transcription initiation to becoming hotspots of elongating molecules.
3/n We used single-molecule tracking and lattice light-sheet microscopy in live Drosophila embryos to ask how clusters change during zygotic genome activation (when there is a ~3x increase in transcription) and how clusters influence transcription levels.
February 12, 2025 at 11:36 AM
3/n We used single-molecule tracking and lattice light-sheet microscopy in live Drosophila embryos to ask how clusters change during zygotic genome activation (when there is a ~3x increase in transcription) and how clusters influence transcription levels.
1/n Excited to share our new preprint! Using live imaging in Drosophila embryos, we show that RNA Pol II clusters switch from sites of initiation to elongation during zygotic genome activation and that they are stably associated with an active gene: www.biorxiv.org/content/10.1...
February 12, 2025 at 11:36 AM
1/n Excited to share our new preprint! Using live imaging in Drosophila embryos, we show that RNA Pol II clusters switch from sites of initiation to elongation during zygotic genome activation and that they are stably associated with an active gene: www.biorxiv.org/content/10.1...
Big congratulations to our MD/PHD student @driaadigun.bsky.social on receiving a F31 award from NIGMS to "Illuminate the Molecular Mechanisms of Replication and Transcription Coordination" during zygotic genome activation and support her training.

January 17, 2025 at 2:10 PM
Big congratulations to our MD/PHD student @driaadigun.bsky.social on receiving a F31 award from NIGMS to "Illuminate the Molecular Mechanisms of Replication and Transcription Coordination" during zygotic genome activation and support her training.
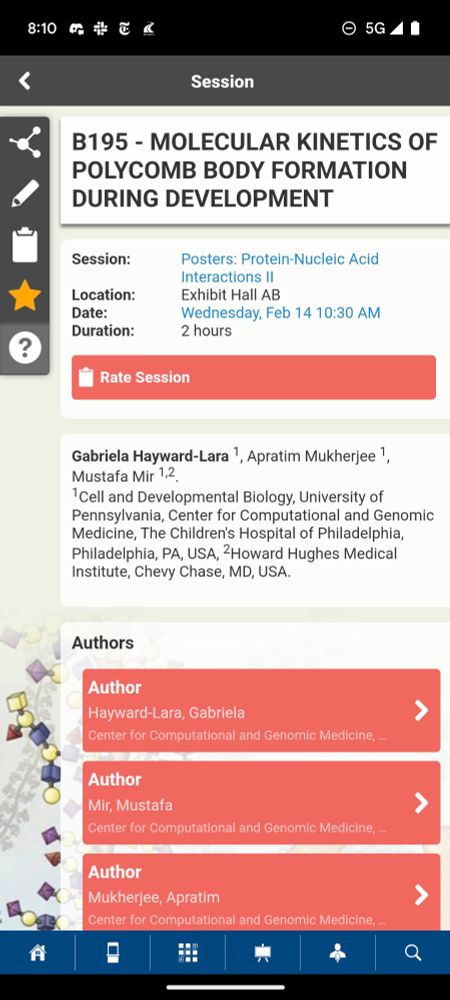
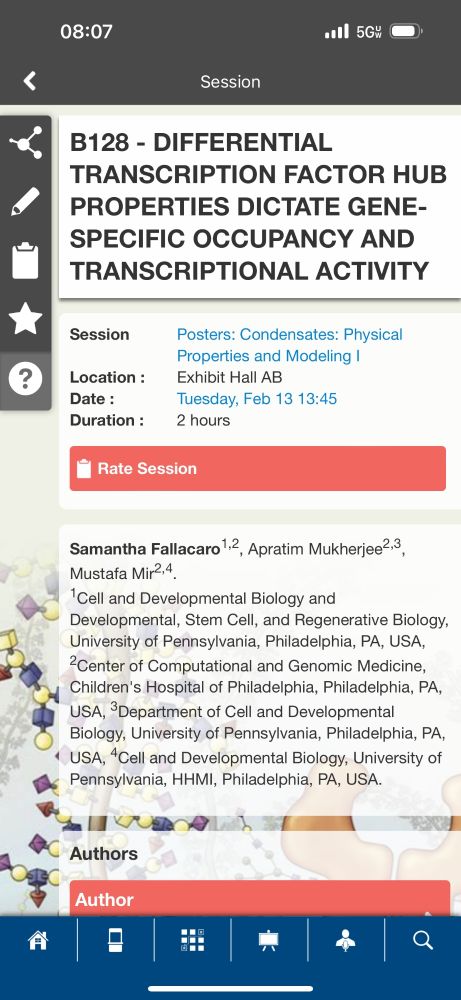
#BPS2024 check out our lab's presentations & pls reach out if you want to chat! Posters:B128 (@littlescholar_ ) txn activator hubs, B195(@GHaywardLara ) repressive bodies. Talks: @mukherja PolII kinetics(13th,5:30 p 201AB), myself, on how hubs find their targets (14th,9:30a BRA)

February 10, 2024 at 1:53 PM
#BPS2024 check out our lab's presentations & pls reach out if you want to chat! Posters:B128 (@littlescholar_ ) txn activator hubs, B195(@GHaywardLara ) repressive bodies. Talks: @mukherja PolII kinetics(13th,5:30 p 201AB), myself, on how hubs find their targets (14th,9:30a BRA)
Happy holidays from our lab to yours!



December 22, 2023 at 1:57 PM
Happy holidays from our lab to yours!
Happy Halloween from the lab, all whom decided to dress like their very scary PI this year 🎃

October 31, 2023 at 6:45 PM
Happy Halloween from the lab, all whom decided to dress like their very scary PI this year 🎃

