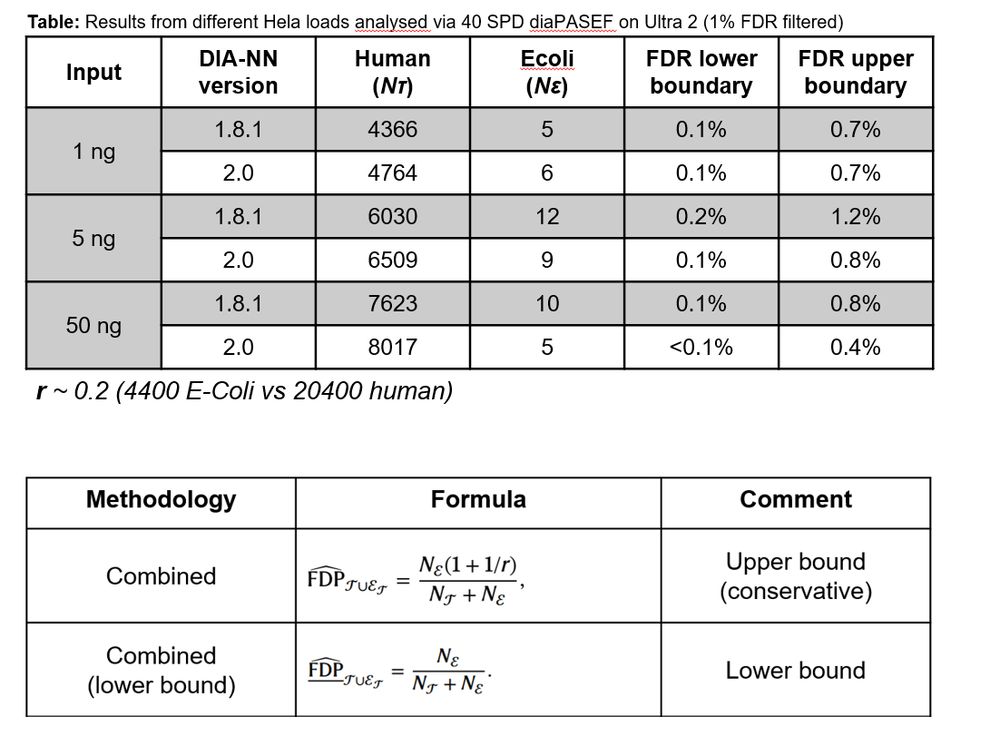

Anyone know what’s going on here? 🤔
#proteomics #bioinformatics @pwilmarth.bsky.social il

Anyone know what’s going on here? 🤔
#proteomics #bioinformatics @pwilmarth.bsky.social il

This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...

This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...


Have tried Fragpipe22? You can do ion mining from DIA data …
Have tried Fragpipe22? You can do ion mining from DIA data …