Protein evolution, metagenomics, AI/ML/DL
Website https://miangoaren.github.io/
Protein sequence-to-structure learning Is this the end(-to-end revolution)
onlinelibrary.wiley.com/doi/abs/10.1...

Protein sequence-to-structure learning Is this the end(-to-end revolution)
onlinelibrary.wiley.com/doi/abs/10.1...
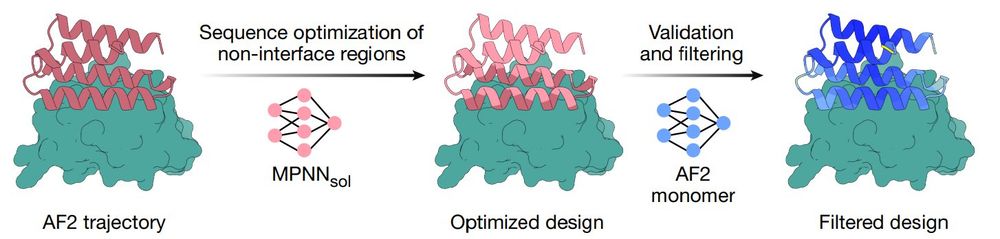
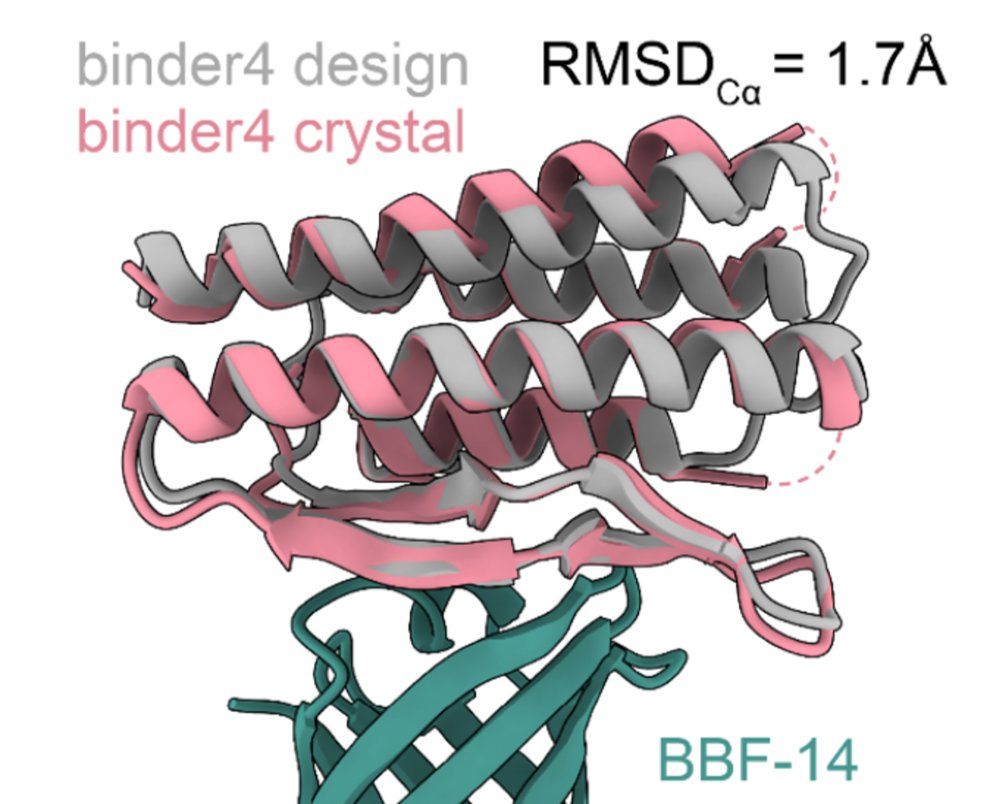
BindCraft
nature.com/articles/s41...
A recent conference about BindCraft
youtube.com/watch?v=qQih...
Boltzdesign1
www.biorxiv.org/content/10.1...
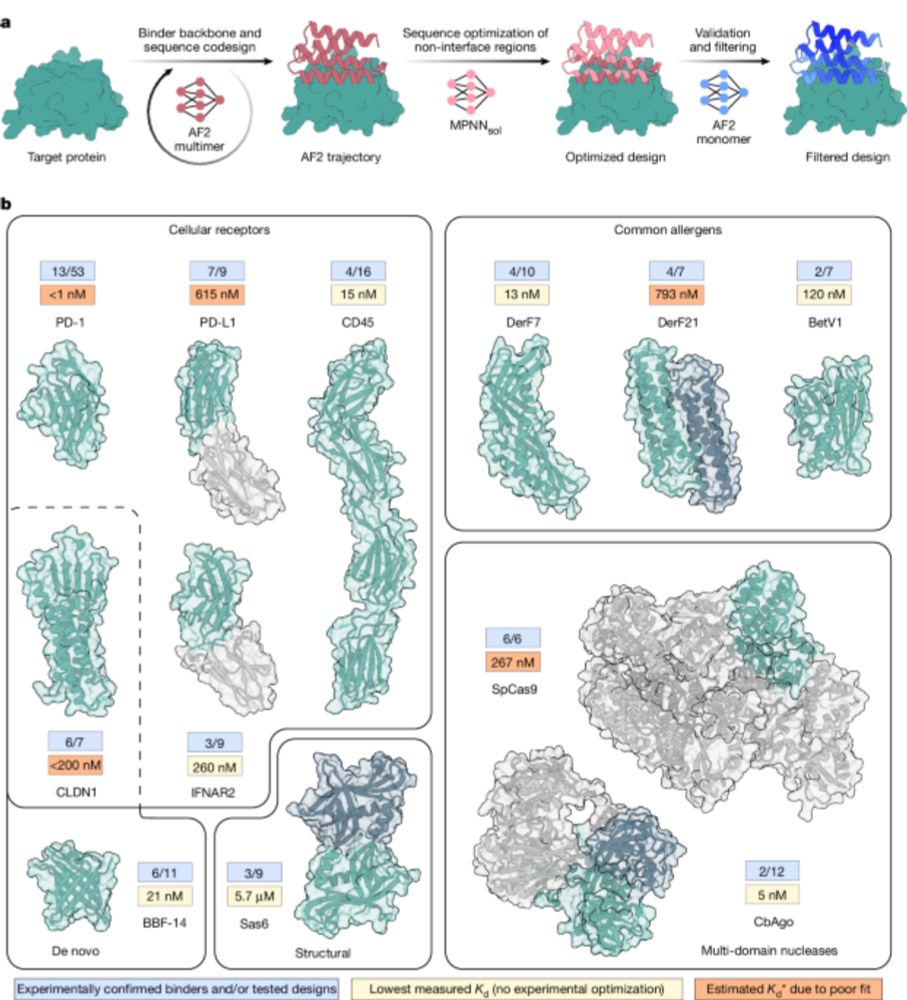
BindCraft
nature.com/articles/s41...
A recent conference about BindCraft
youtube.com/watch?v=qQih...
Boltzdesign1
www.biorxiv.org/content/10.1...
Congrats to all the authors!
Congrats to all the authors!

adaptyvbio.com/blog/po104

adaptyvbio.com/blog/po104
*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies

*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies