Protein evolution, metagenomics, AI/ML/DL
Website https://miangoaren.github.io/



Congrats to all the authors!
Congrats to all the authors!

*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies

*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies
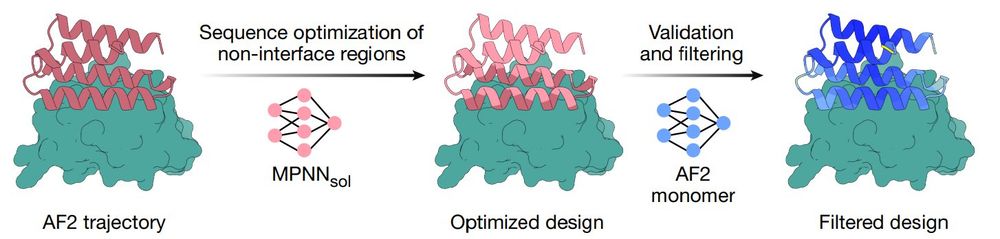





@nature.com , one of the most famous AIs in biology for designing protein–protein interactions (PPI). In my opinion. Bindcraft represents one of the most important advances in the post–AlphaFold2 era.

@nature.com , one of the most famous AIs in biology for designing protein–protein interactions (PPI). In my opinion. Bindcraft represents one of the most important advances in the post–AlphaFold2 era.
Img source (2020)
pubmed.ncbi.nlm.nih.gov/32302382/
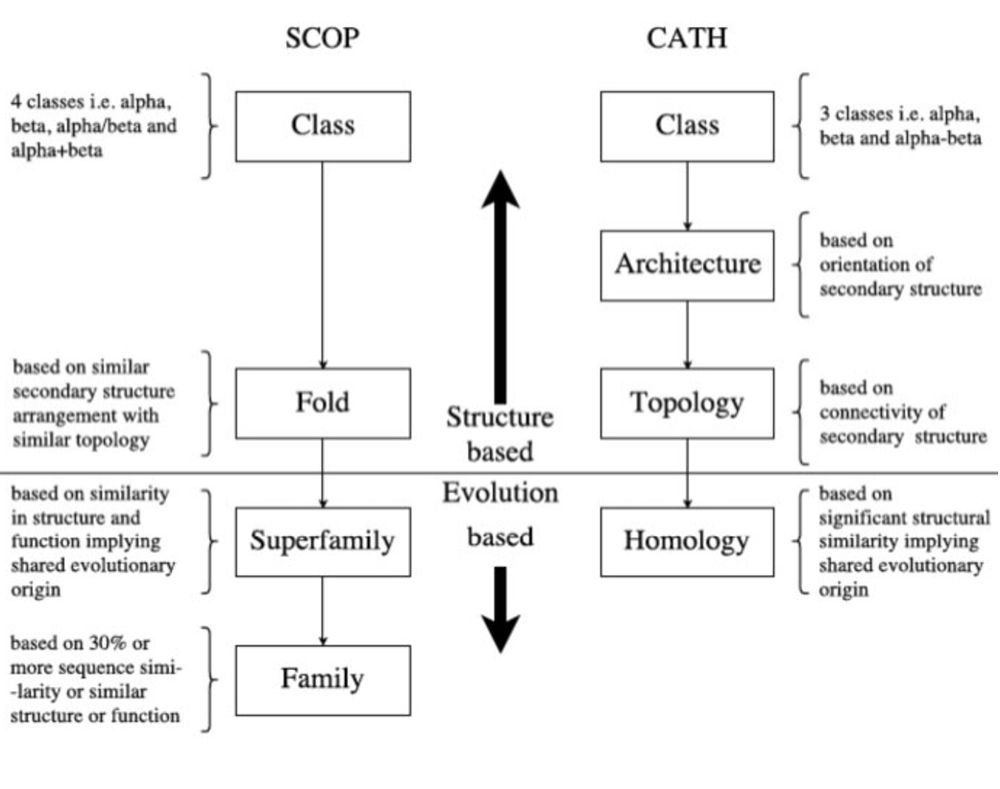
Img source (2020)
pubmed.ncbi.nlm.nih.gov/32302382/
Sensitive protein alignments at tree-of-life scale using DIAMOND
www.nature.com/articles/s41...

Sensitive protein alignments at tree-of-life scale using DIAMOND
www.nature.com/articles/s41...
bsky.app/profile/bris...
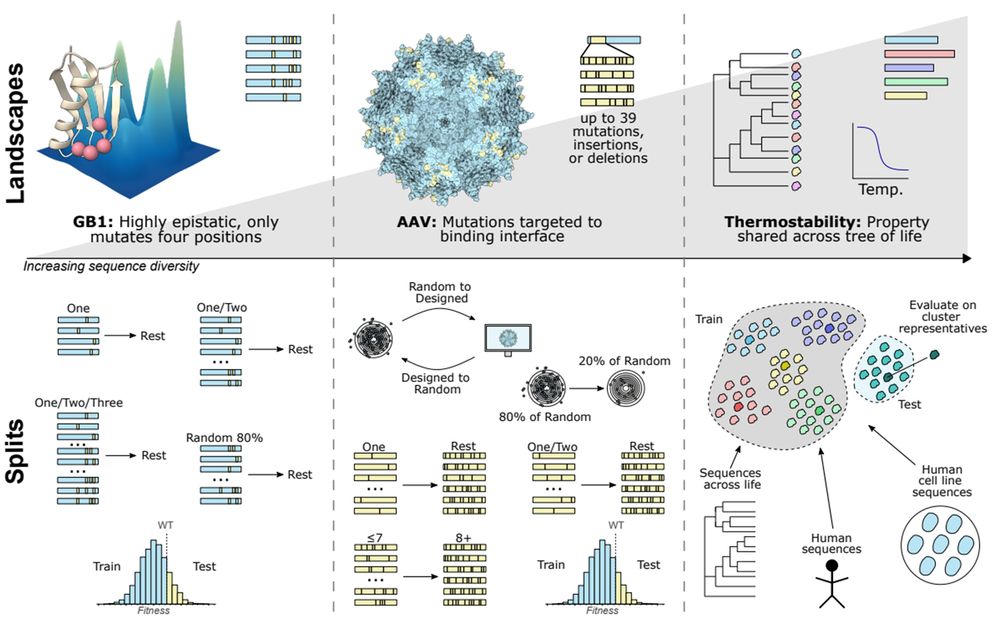
bsky.app/profile/bris...
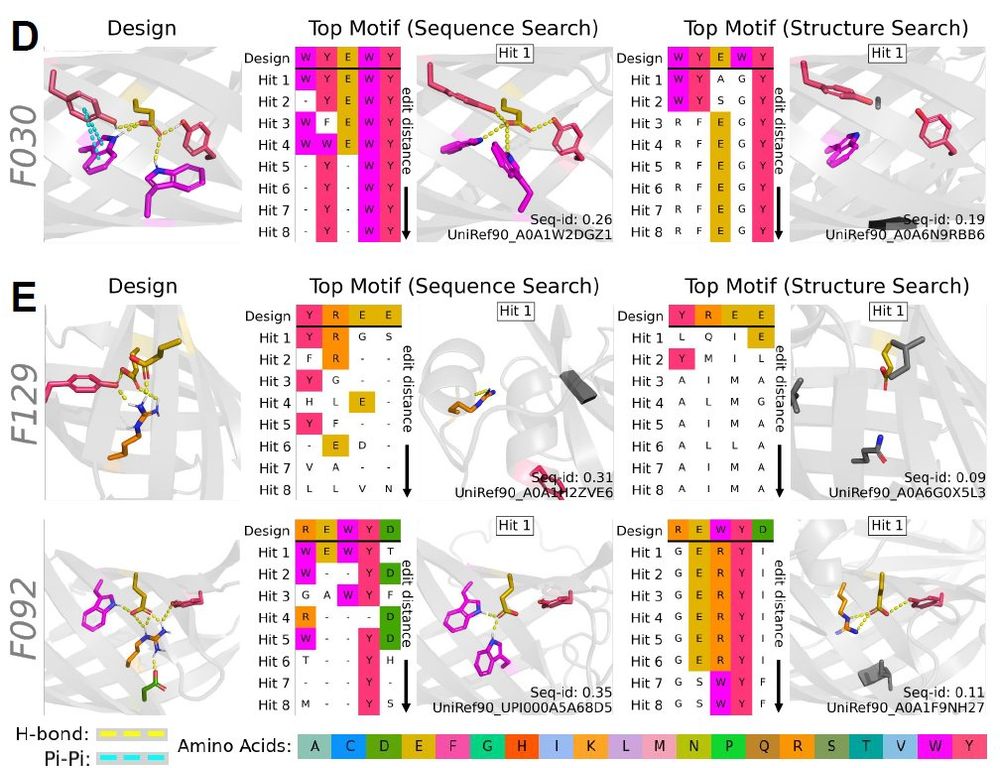


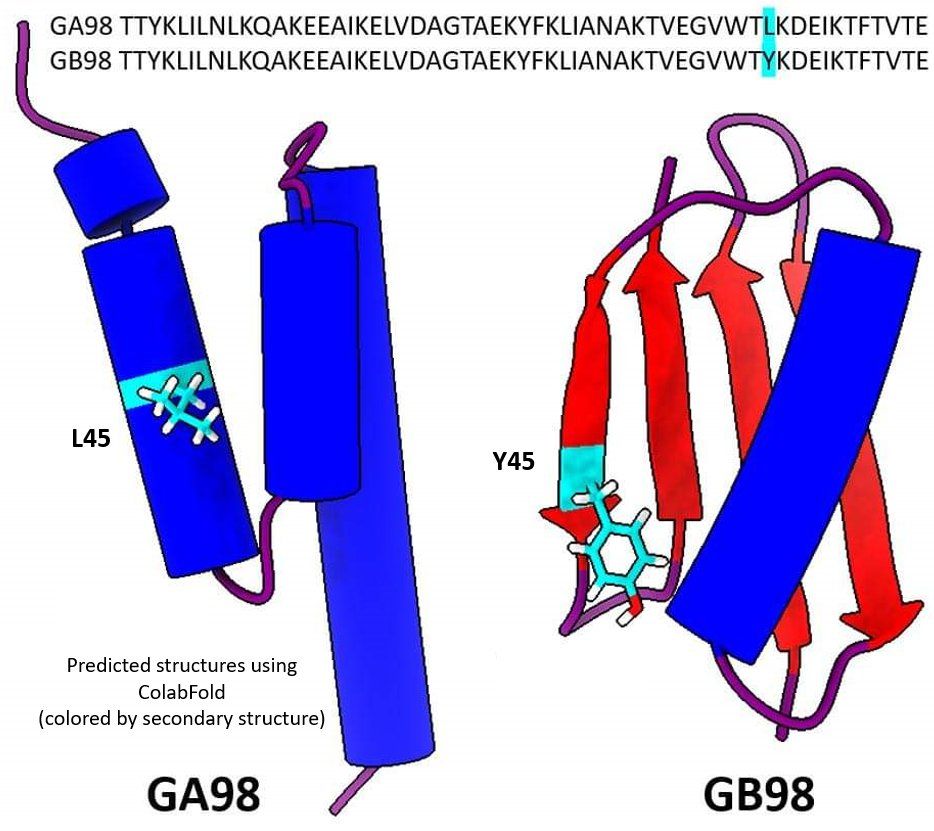
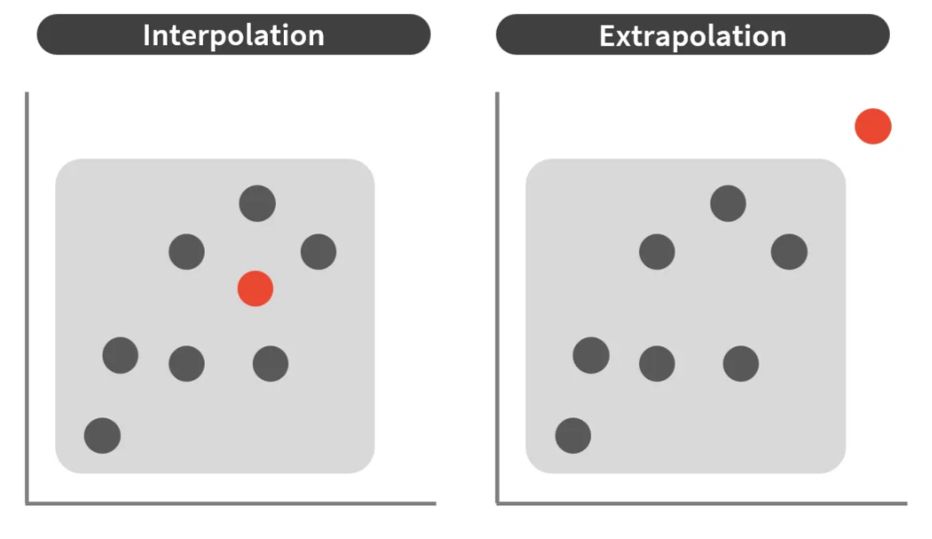
1 historical contingency (i.e., today’s proteins are not truly representative of the full diversity).
2 Proteins are not IID (due to factors like superfolds and shared motifs between non-homologous proteins)

1 historical contingency (i.e., today’s proteins are not truly representative of the full diversity).
2 Proteins are not IID (due to factors like superfolds and shared motifs between non-homologous proteins)


youtu.be/P_fHJIYENdI?...
But also (and mainly), in this answer by Ilya Sutskever at the last NeurIPS about the generalization capabilities of reasoning models
youtu.be/1yvBqasHLZs?...


youtu.be/P_fHJIYENdI?...
But also (and mainly), in this answer by Ilya Sutskever at the last NeurIPS about the generalization capabilities of reasoning models
youtu.be/1yvBqasHLZs?...