Max Fürst
@maxfus.bsky.social
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
I set myself a reminder to check this 1 year later & turns out I was mostly wrong🥲
Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?
Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?

November 5, 2025 at 4:06 PM
I set myself a reminder to check this 1 year later & turns out I was mostly wrong🥲
Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?
Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?
wouldn't it make much more sense to plot this relative to normal distribution found across the proteome?
Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...
Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...

October 27, 2025 at 5:14 PM
wouldn't it make much more sense to plot this relative to normal distribution found across the proteome?
Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...
Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...
Un-frigging-believable. I have waited for something like this for so long!
October 5, 2025 at 8:01 AM
Un-frigging-believable. I have waited for something like this for so long!
I kept telling people this in discussions, until someone German insisted so I looked it up and it's indeed not that clear.
Here formatted in a similar way from source www.bka.de/DE/AktuelleI...
Here formatted in a similar way from source www.bka.de/DE/AktuelleI...

August 8, 2025 at 9:43 PM
I kept telling people this in discussions, until someone German insisted so I looked it up and it's indeed not that clear.
Here formatted in a similar way from source www.bka.de/DE/AktuelleI...
Here formatted in a similar way from source www.bka.de/DE/AktuelleI...
Now that OpenCRISPR is in nature and rekindled the 'what's-a-novel-sequence' debate, I'm happy to share an app to check this, which I built for fun some time ago.
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
August 1, 2025 at 11:50 AM
Now that OpenCRISPR is in nature and rekindled the 'what's-a-novel-sequence' debate, I'm happy to share an app to check this, which I built for fun some time ago.
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
Fixed that for you!
Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️
Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️


June 2, 2025 at 8:16 AM
Fixed that for you!
Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️
Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️
As a final application example, we demonstrate protein immobiliziation. Commercial azido agarose reacted happily with ADD-tagged GFP, and we got a brightly shining resin after reacting the protein on-column with ApbE!

May 20, 2025 at 12:30 PM
As a final application example, we demonstrate protein immobiliziation. Commercial azido agarose reacted happily with ADD-tagged GFP, and we got a brightly shining resin after reacting the protein on-column with ApbE!
Conjugating commercial click-modified DNA oligos also worked at the first shot without optimization, although at relatively low conjugation yields. We’re not so experienced with the chemistry, but apparently Cu catalysis isn’t great for DNA either, so probably switching to SPAAC would work better.

May 20, 2025 at 12:30 PM
Conjugating commercial click-modified DNA oligos also worked at the first shot without optimization, although at relatively low conjugation yields. We’re not so experienced with the chemistry, but apparently Cu catalysis isn’t great for DNA either, so probably switching to SPAAC would work better.
As expected, biomolecule conjugation needs more care: we saw moderate conversion yields in protein-protein conjugations (N-N/C-C/branched). We did not optimize our proof of concept—safe to assume that it could easily be pushed by varying position, adding linkers, optimized reaction conditions, etc.

May 20, 2025 at 12:30 PM
As expected, biomolecule conjugation needs more care: we saw moderate conversion yields in protein-protein conjugations (N-N/C-C/branched). We did not optimize our proof of concept—safe to assume that it could easily be pushed by varying position, adding linkers, optimized reaction conditions, etc.
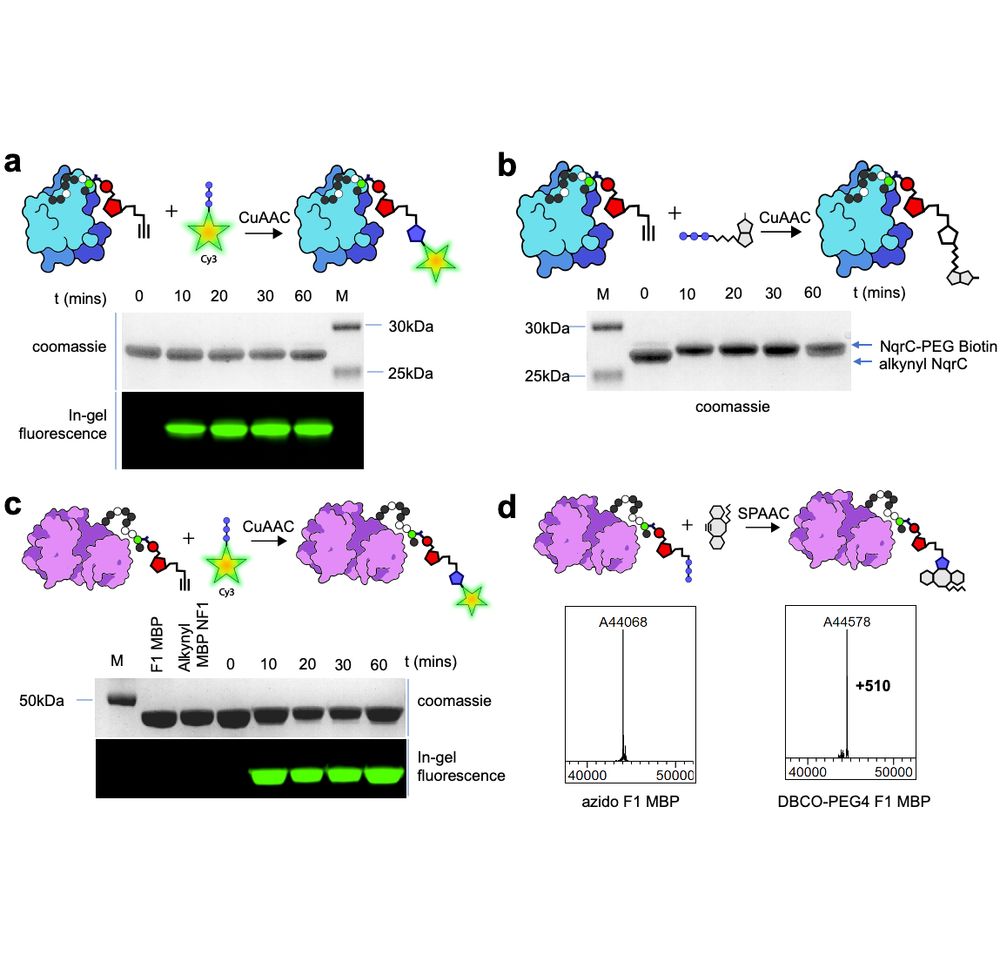
Small molecules were readily conjugated to all our example proteins: we show attachment of fluorophores & biotin, and establish that both classic copper-catalyzed, as well as @carolynbertozzi.bskyverified.social's copper-free SPAAC reaction are compatible with ADD tags & result in full labeling.
May 20, 2025 at 12:30 PM
Small molecules were readily conjugated to all our example proteins: we show attachment of fluorophores & biotin, and establish that both classic copper-catalyzed, as well as @carolynbertozzi.bskyverified.social's copper-free SPAAC reaction are compatible with ADD tags & result in full labeling.
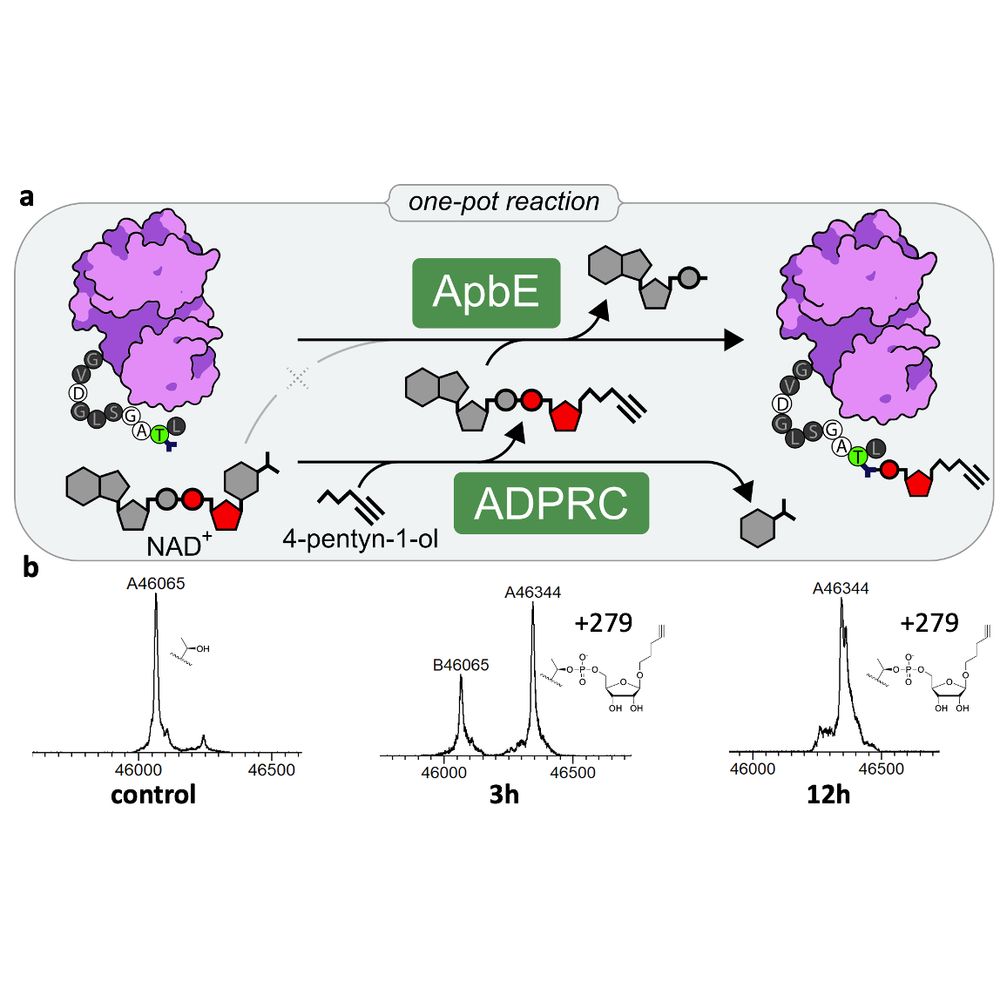
Because ADPRC works on NAD+, not NADPH, while ApbE only accepts NADPH and ignores NAD+, we can even run the two enzymatic steps (substrate generation & protein modification) in one pot—just add the two enzymes with the precursors NAD+ and the click alkanol to your target protein!
May 20, 2025 at 12:30 PM
Because ADPRC works on NAD+, not NADPH, while ApbE only accepts NADPH and ignores NAD+, we can even run the two enzymatic steps (substrate generation & protein modification) in one pot—just add the two enzymes with the precursors NAD+ and the click alkanol to your target protein!
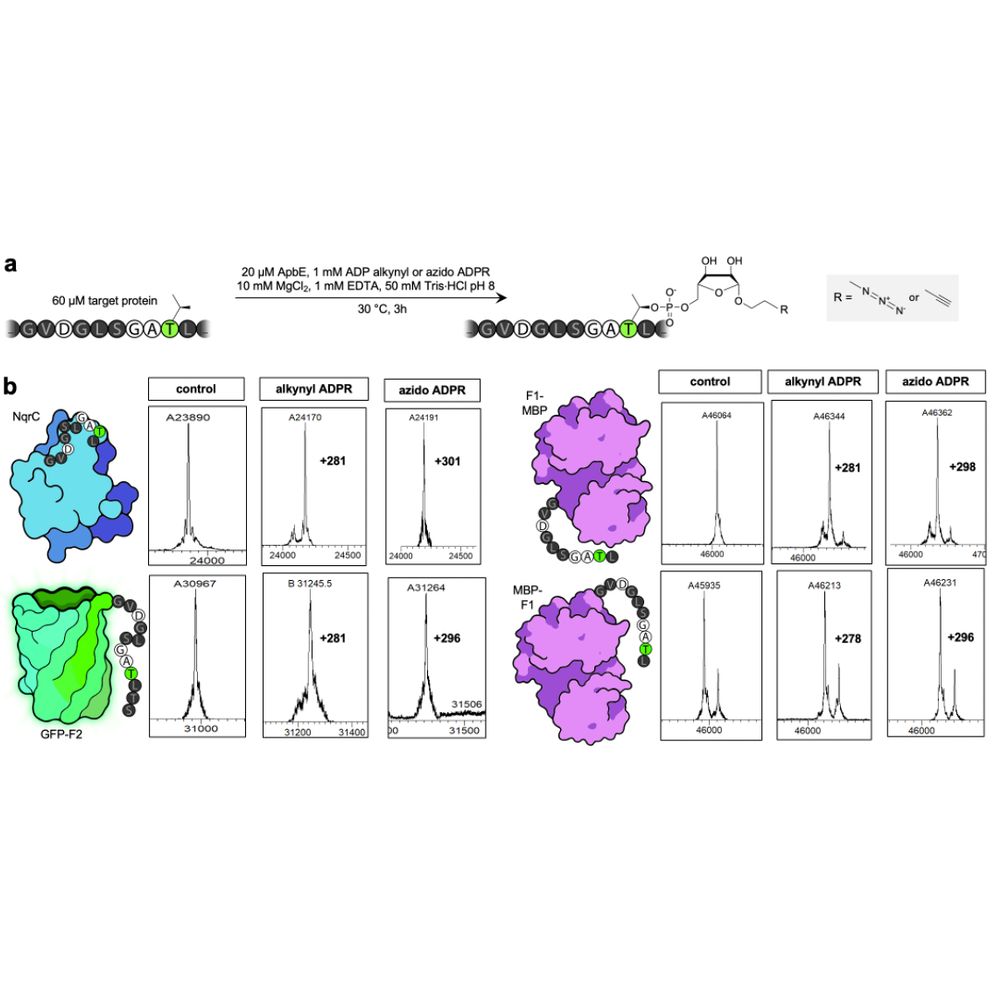
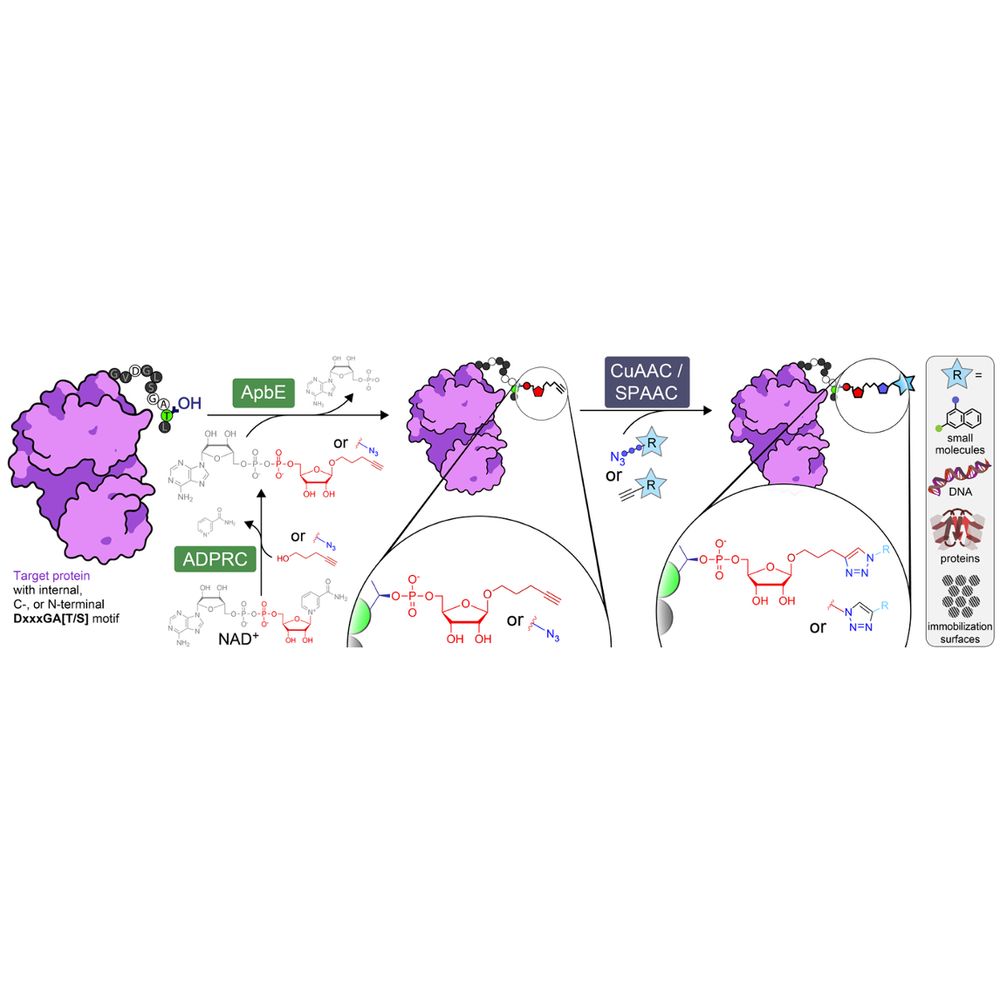
Turns out ApbE is happily accepting this substrate, allowing us to label a variety of proteins at N or C terminus, as well as internally, with either alkynyl or azido ribose phosphate moieties, yielding typically full conversion after a few hours of reaction, as determined by MS.
May 20, 2025 at 12:30 PM
Turns out ApbE is happily accepting this substrate, allowing us to label a variety of proteins at N or C terminus, as well as internally, with either alkynyl or azido ribose phosphate moieties, yielding typically full conversion after a few hours of reaction, as determined by MS.
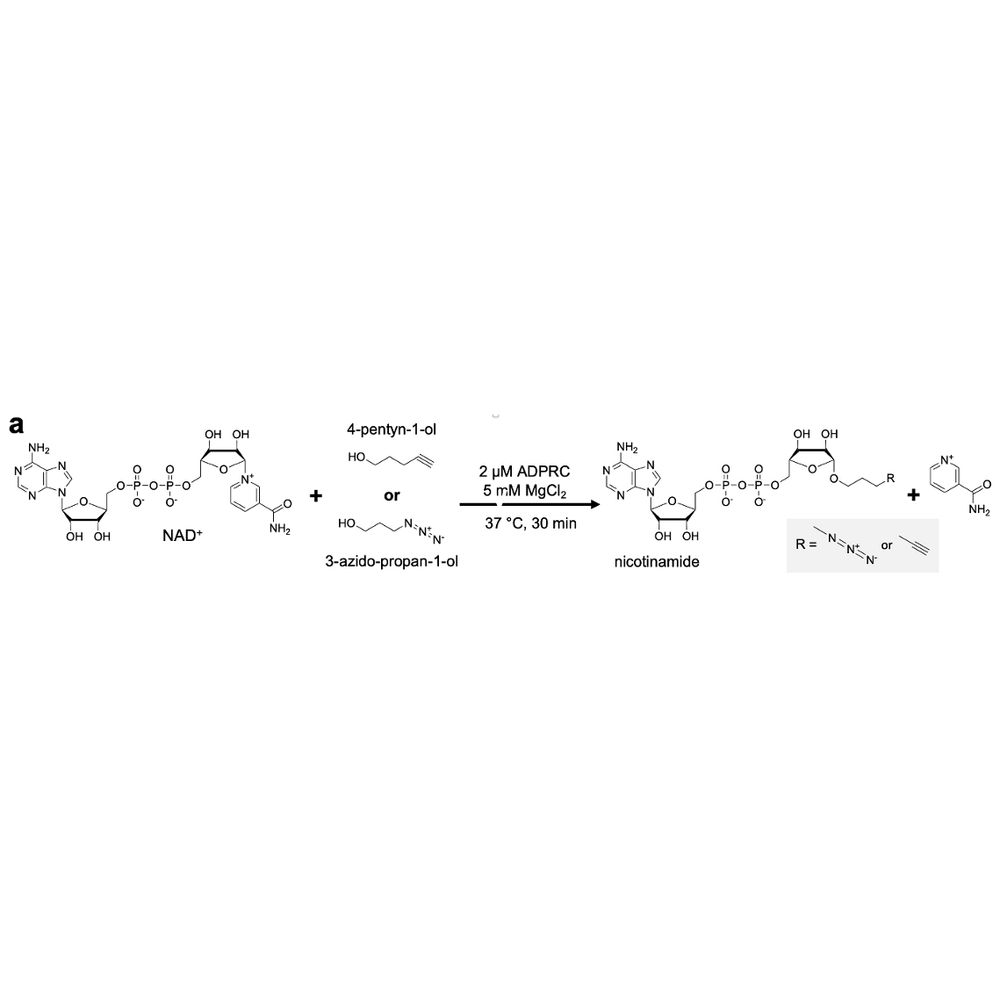
Given the enzyme’s reported promiscuity toward NADH, we hypothesised that it tolerates synthetic substrates with similar structure. Courtesy to Jäschke lab developing NAD capture seq, we knew that a commercial enzyme—ADPRC—can generate a click dinucleotide from cheap small molecule alcohols & NAD+
May 20, 2025 at 12:30 PM
Given the enzyme’s reported promiscuity toward NADH, we hypothesised that it tolerates synthetic substrates with similar structure. Courtesy to Jäschke lab developing NAD capture seq, we knew that a commercial enzyme—ADPRC—can generate a click dinucleotide from cheap small molecule alcohols & NAD+
🚨 preprint 2️⃣ this month: our (purely experimental🧪) venture into #ChemBio
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
May 20, 2025 at 12:30 PM
🚨 preprint 2️⃣ this month: our (purely experimental🧪) venture into #ChemBio
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
BoostMut's output is a table of scores on various metrics & we provide a web app to explore.
While the default score is simple mean, we trained some lightweight ML models to assigned weights. They did a bit better, we're currently still exploring!
While the default score is simple mean, we trained some lightweight ML models to assigned weights. They did a bit better, we're currently still exploring!

May 7, 2025 at 5:04 PM
BoostMut's output is a table of scores on various metrics & we provide a web app to explore.
While the default score is simple mean, we trained some lightweight ML models to assigned weights. They did a bit better, we're currently still exploring!
While the default score is simple mean, we trained some lightweight ML models to assigned weights. They did a bit better, we're currently still exploring!
We also ran fresh MDs on large literature datasets that used ML predictors: again, small but consistent ROC AUC gains.
Finally, we tested BoostMut's top mutations in the lab, and: even higher success rate than manual selected mutations!
Finally, we tested BoostMut's top mutations in the lab, and: even higher success rate than manual selected mutations!

May 7, 2025 at 5:04 PM
We also ran fresh MDs on large literature datasets that used ML predictors: again, small but consistent ROC AUC gains.
Finally, we tested BoostMut's top mutations in the lab, and: even higher success rate than manual selected mutations!
Finally, we tested BoostMut's top mutations in the lab, and: even higher success rate than manual selected mutations!
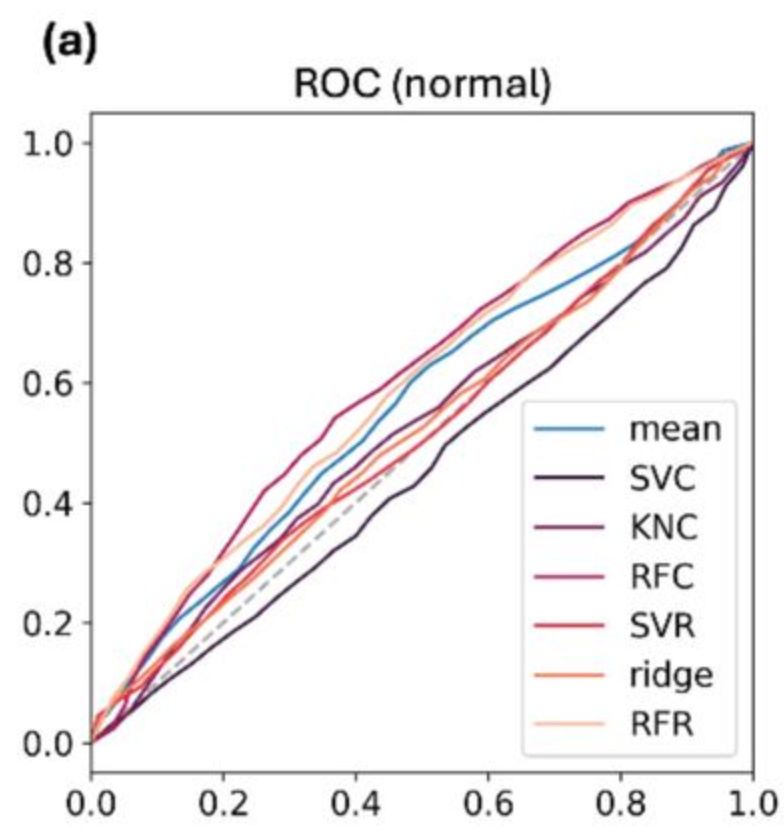
We made a huge effort to benchmark thoroughly, mostly via ROC analysis. First on old FRESCO projects: predictions & MDs were already there. BoostMut's top hits overlapped manual picks, though not 1:1. But: success rates were as good or better.


May 7, 2025 at 5:04 PM
We made a huge effort to benchmark thoroughly, mostly via ROC analysis. First on old FRESCO projects: predictions & MDs were already there. BoostMut's top hits overlapped manual picks, though not 1:1. But: success rates were as good or better.
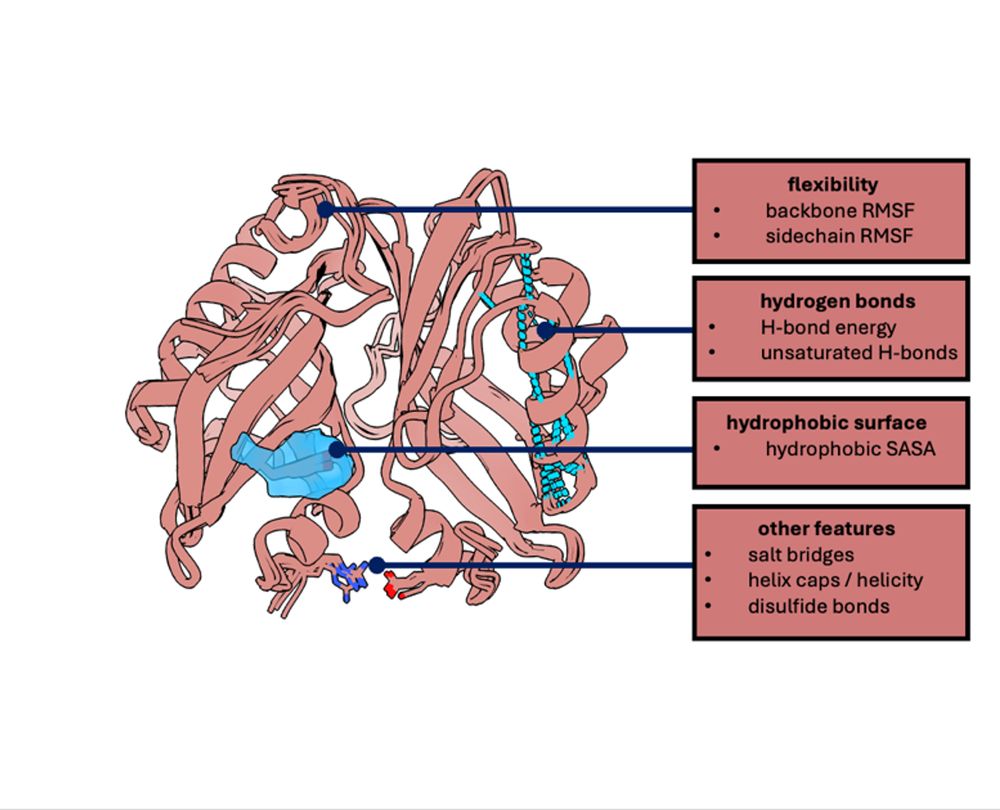
Built on Python’s MDAnalysis, BoostMut formalizes key stability checks: H-bonds ↑, hydrophobic exposure ↓, flexibility ↓, etc.
Some metrics were tricky, e.g. what’s “normal” for sidechain flex? We simulated high-res PDBs, binned by surface exposure, & fit a monotonic curve to not penalize rigidity.
Some metrics were tricky, e.g. what’s “normal” for sidechain flex? We simulated high-res PDBs, binned by surface exposure, & fit a monotonic curve to not penalize rigidity.

May 7, 2025 at 5:04 PM
Built on Python’s MDAnalysis, BoostMut formalizes key stability checks: H-bonds ↑, hydrophobic exposure ↓, flexibility ↓, etc.
Some metrics were tricky, e.g. what’s “normal” for sidechain flex? We simulated high-res PDBs, binned by surface exposure, & fit a monotonic curve to not penalize rigidity.
Some metrics were tricky, e.g. what’s “normal” for sidechain flex? We simulated high-res PDBs, binned by surface exposure, & fit a monotonic curve to not penalize rigidity.
Given that this is in the same realm as what we achieved in previous FRESCO projects, e.g. when we engineered an alcohol dehydrogenase to boiling point stability (5 years ago) we consider the stabilizing mutation prediction problem as not solved at this point in time
elifesciences.org/articles/54639
elifesciences.org/articles/54639

May 7, 2025 at 5:04 PM
Given that this is in the same realm as what we achieved in previous FRESCO projects, e.g. when we engineered an alcohol dehydrogenase to boiling point stability (5 years ago) we consider the stabilizing mutation prediction problem as not solved at this point in time
elifesciences.org/articles/54639
elifesciences.org/articles/54639
While primary prediction could these days be ML-based, we haven't really given up on energy-based ones, due to very similar hit rates. Take e.g. PRIME, a recent "SOTA" model. Great paper, as (unlike most new fancy model papers) they actually put their tool to the test in the lab, but hit rate = 40%

May 7, 2025 at 5:04 PM
While primary prediction could these days be ML-based, we haven't really given up on energy-based ones, due to very similar hit rates. Take e.g. PRIME, a recent "SOTA" model. Great paper, as (unlike most new fancy model papers) they actually put their tool to the test in the lab, but hit rate = 40%
We've engineered thermostable proteins in Groningen for many years, traditionally using our FRESCO pipeline, which uses MD screens to sort out false positives from thermostability predictors such as FoldX. This has been highly successful, stabilizing proteins on average by +20 °C with ~8 mutations.

May 7, 2025 at 5:04 PM
We've engineered thermostable proteins in Groningen for many years, traditionally using our FRESCO pipeline, which uses MD screens to sort out false positives from thermostability predictors such as FoldX. This has been highly successful, stabilizing proteins on average by +20 °C with ~8 mutations.
Preprint from the lab🚨
Have you ever engineered proteins to be more stable and were unhappy about your predictor's success rate? We got you covered with BoostMut!
Great work led by @kerlenkorbeld.bsky.social now online at www.biorxiv.org/content/10.1...
A thread 🧵
Have you ever engineered proteins to be more stable and were unhappy about your predictor's success rate? We got you covered with BoostMut!
Great work led by @kerlenkorbeld.bsky.social now online at www.biorxiv.org/content/10.1...
A thread 🧵
May 7, 2025 at 5:04 PM
Preprint from the lab🚨
Have you ever engineered proteins to be more stable and were unhappy about your predictor's success rate? We got you covered with BoostMut!
Great work led by @kerlenkorbeld.bsky.social now online at www.biorxiv.org/content/10.1...
A thread 🧵
Have you ever engineered proteins to be more stable and were unhappy about your predictor's success rate? We got you covered with BoostMut!
Great work led by @kerlenkorbeld.bsky.social now online at www.biorxiv.org/content/10.1...
A thread 🧵
Covid Spike protein, PDB 6CRZ

May 3, 2025 at 12:40 PM
Covid Spike protein, PDB 6CRZ
copy your favorite outcome to Adobe Illustrator and use Image Trace with the preset 3 or 6 Colors. Expand, and adjust colors to your liking


May 3, 2025 at 12:40 PM
copy your favorite outcome to Adobe Illustrator and use Image Trace with the preset 3 or 6 Colors. Expand, and adjust colors to your liking


