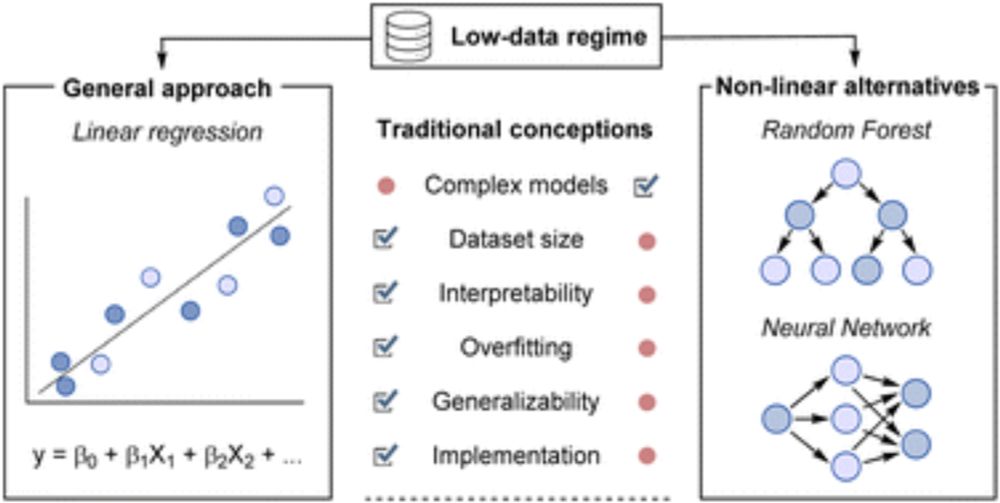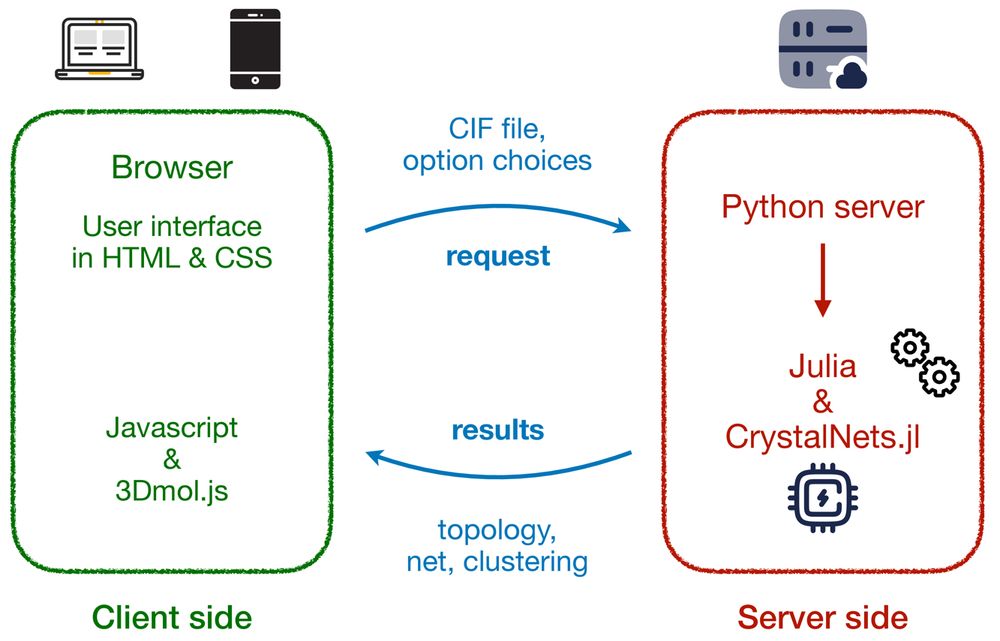
Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....

Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....
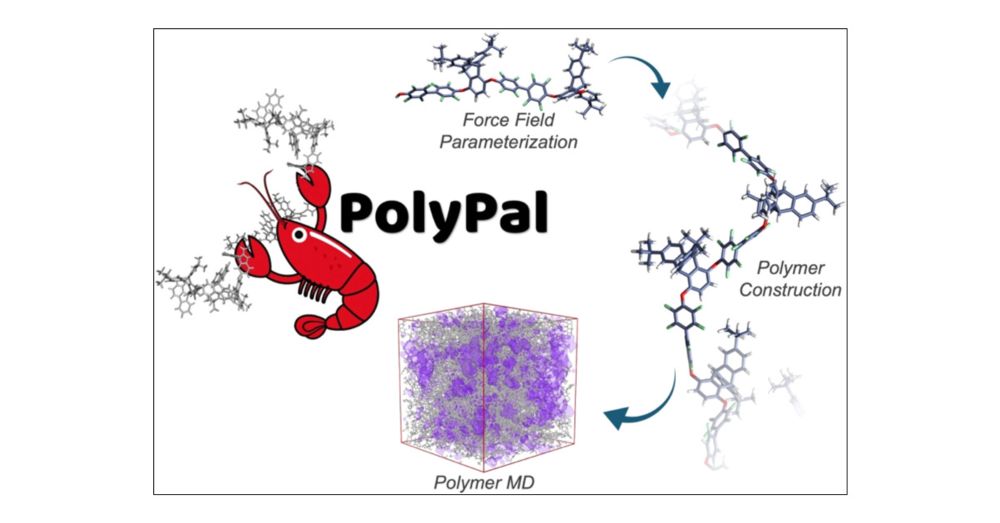
doi.org/10.1021/acs....

doi.org/10.1021/acs....
doi.org/10.1021/acs....


#WATOC #CompChemSky #ChemSky
www.watoc2025.no
#WATOC #CompChemSky #ChemSky
www.watoc2025.no

pubs.acs.org/doi/10.1021/...
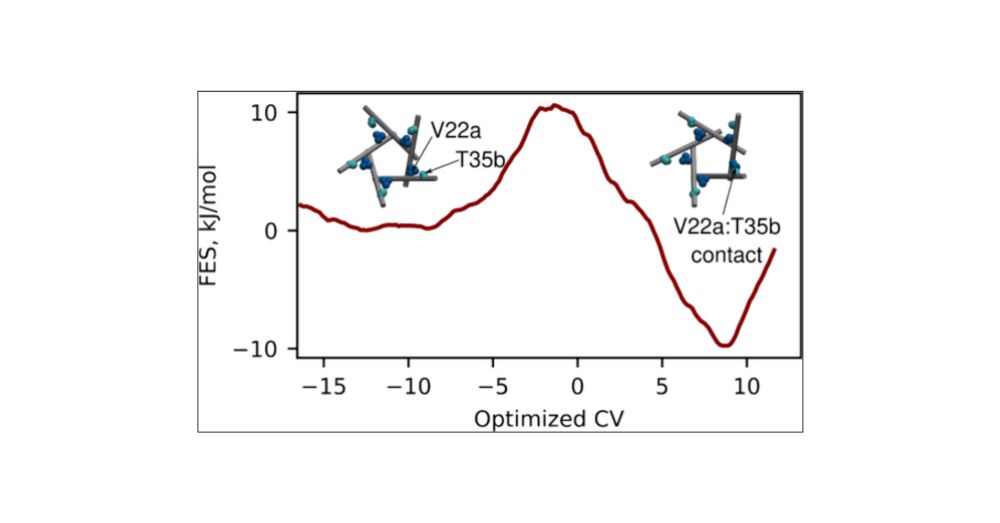
pubs.acs.org/doi/10.1021/...
#compchemsky #chemsky
#compchemsky #chemsky
Also check out our recent webinars on each:
🎬 Whole cell simulation with Martini ▶️ youtu.be/fvFaPgSoM90
🎬 Visual analysis of #moleculardynamics with VIAMD ▶️ youtu.be/wVENzcx0XmQ
Also check out our recent webinars on each:
🎬 Whole cell simulation with Martini ▶️ youtu.be/fvFaPgSoM90
🎬 Visual analysis of #moleculardynamics with VIAMD ▶️ youtu.be/wVENzcx0XmQ
: "The Q-AMOEBA (CF) Polarizable Potential".
Enhanced version of the Q-AMOEBA polarizable model integrating a geometry-dependent charge flux term while designed for an explicit treatment of nuclear quantum effects.
pubs.acs.org/doi/10.1021/...
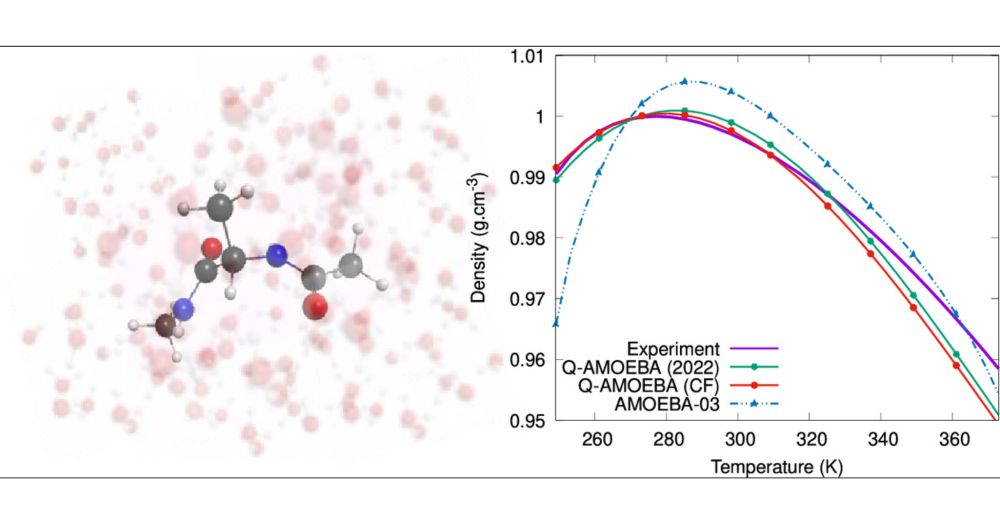
: "The Q-AMOEBA (CF) Polarizable Potential".
Enhanced version of the Q-AMOEBA polarizable model integrating a geometry-dependent charge flux term while designed for an explicit treatment of nuclear quantum effects.
pubs.acs.org/doi/10.1021/...
doi.org/10.1093/bioi...
VTX is open source and freely accessible for non commercial use! github.com/VTX-Molecula...
Builds available at vtx.drugdesign.fr
doi.org/10.1093/bioi...
VTX is open source and freely accessible for non commercial use! github.com/VTX-Molecula...
Builds available at vtx.drugdesign.fr
Join the VeloxChem on LUMI Workshop — 26–27 May 2025 in Espoo, Finland!
💡 Learn to:
Run VeloxChem via Jupyter
Scale jobs on LUMI's GPUs with Slurm
Analyze results with VIAMD
Info + signup 👉 lumi-supercomputer.eu/events/velox...
lumi-supercomputer.eu/events/velox...