
We define the Protein Dark Energy and use it to localize and quantify the functional evolutionary constraints on natural proteins
arxiv.org/abs/2508.08109

We define the Protein Dark Energy and use it to localize and quantify the functional evolutionary constraints on natural proteins
arxiv.org/abs/2508.08109
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
www.nature.com/articles/s41...
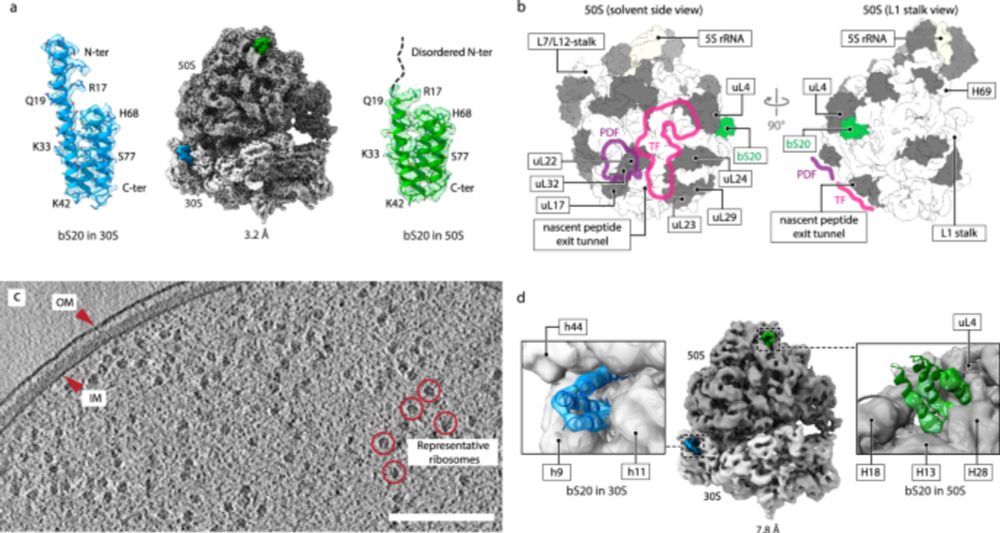
www.nature.com/articles/s41...
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
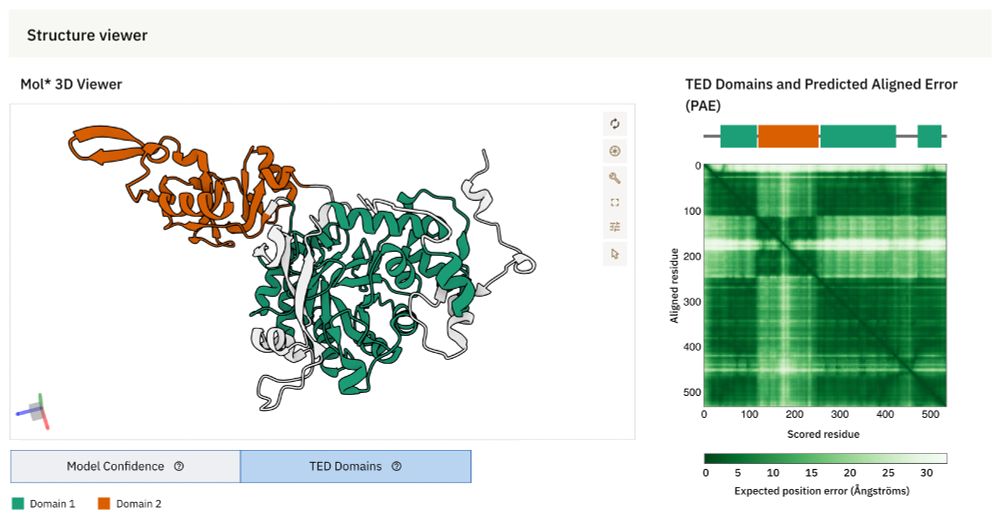
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social

We are thrilled to learn about the experiences, challenges and achievements from our incredible network nodes 👇
#ImagingTheFuture

We are thrilled to learn about the experiences, challenges and achievements from our incredible network nodes 👇
#ImagingTheFuture
#CEBEM #ImagingTheFuture

#CEBEM #ImagingTheFuture
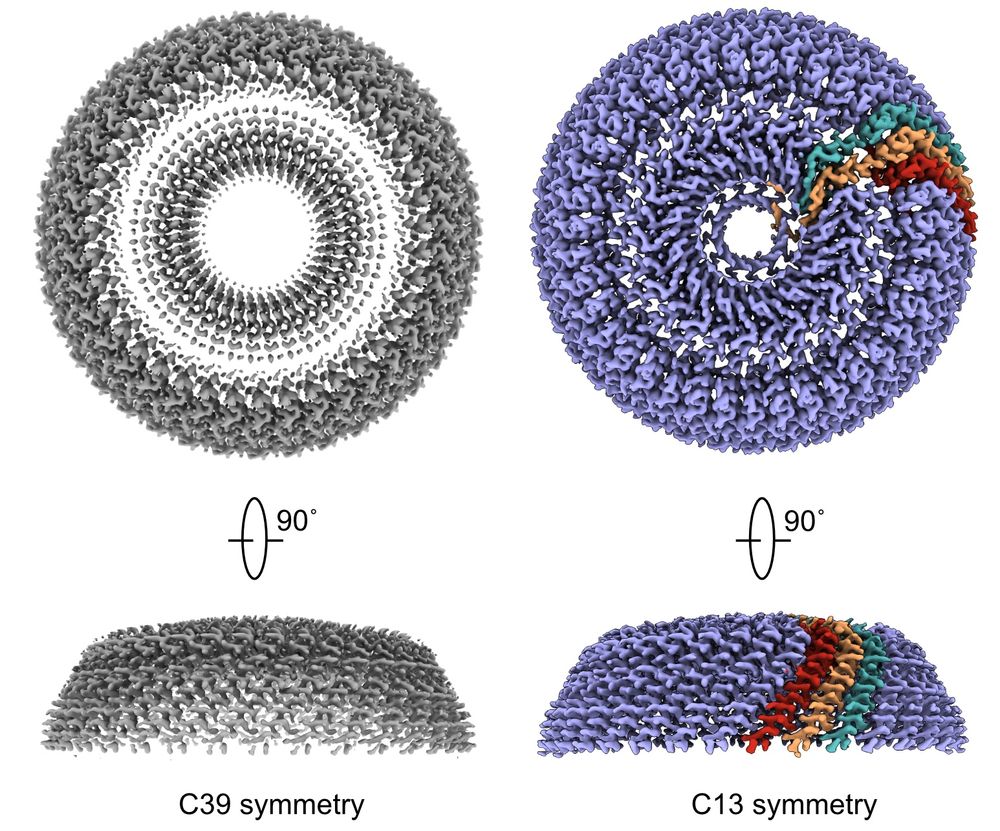
#virology #cryoEM
www.nature.com/articles/s41...

#virology #cryoEM
www.nature.com/articles/s41...

