Matthew A. Clarke
@maclarke.bsky.social
Computational biologist and mechanistic interpretability researcher. For more find me at: https://mclarke1991.github.io & https://www.linkedin.com/in/matthew-alan-clarke/
Huge thanks to co-first authors @charliegbarker.bsky.social & Yuxin Sun for their exceptional efforts on this project, to our wonderful collaborators, and especially to the Jasmin Fisher lab at @ucl.ac.uk Cancer Institute!
The pre-print is open access & available at: www.biorxiv.org/content/10.1...
The pre-print is open access & available at: www.biorxiv.org/content/10.1...

MAGELLAN: Automated Generation of Interpretable Computational Models for Biological Reasoning
Computational models have become essential tools for understanding signalling networks and their non-linear dynamics. However, these models are typically constructed manually using prior knowledge and...
www.biorxiv.org
May 26, 2025 at 9:50 PM
Huge thanks to co-first authors @charliegbarker.bsky.social & Yuxin Sun for their exceptional efforts on this project, to our wonderful collaborators, and especially to the Jasmin Fisher lab at @ucl.ac.uk Cancer Institute!
The pre-print is open access & available at: www.biorxiv.org/content/10.1...
The pre-print is open access & available at: www.biorxiv.org/content/10.1...
This model can then be further edited and interrogated using the BioModelAnalyzer GUI (biomodelanalyzer.org) and command-line tools (github.com/hallba/BioMo...).

May 26, 2025 at 9:50 PM
This model can then be further edited and interrogated using the BioModelAnalyzer GUI (biomodelanalyzer.org) and command-line tools (github.com/hallba/BioMo...).
MAGELLAN allows the generation of a computational model from pathway databases, and the fitting of these models to a user-specified specification, resulting in a transparent and interpretable model.

May 26, 2025 at 9:50 PM
MAGELLAN allows the generation of a computational model from pathway databases, and the fitting of these models to a user-specified specification, resulting in a transparent and interpretable model.
Reposted by Matthew A. Clarke
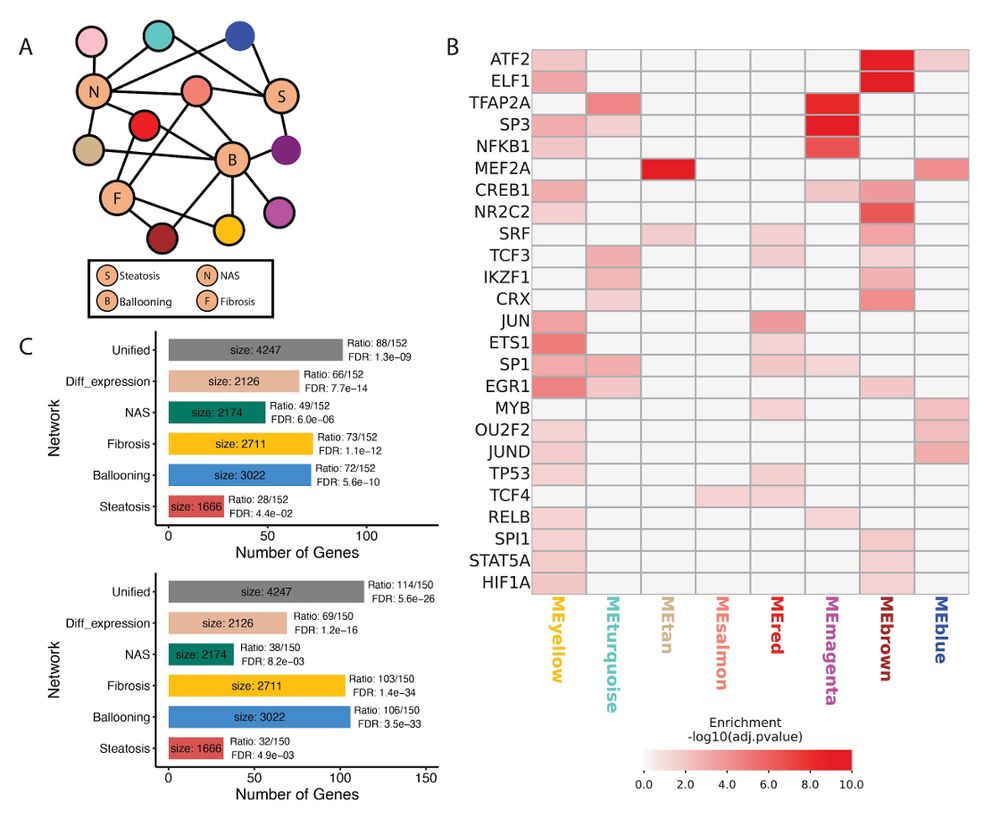
We then used our published method from @charliegbarker.bsky.social (Barker et al, Genome Research, 2022) to create a reference MASLD network by associating gene co-expression modules with histology scores, identifying key transcription factors regulating them and applying network reconstruction.
January 19, 2025 at 11:18 AM
We then used our published method from @charliegbarker.bsky.social (Barker et al, Genome Research, 2022) to create a reference MASLD network by associating gene co-expression modules with histology scores, identifying key transcription factors regulating them and applying network reconstruction.
Special thanks to my co-first authors @charliegbarker.bsky.social and Ashley Nicholls for their incredible work on this project, and to the Jasmin Fisher lab and all our collaborators!

January 12, 2025 at 5:29 PM
Special thanks to my co-first authors @charliegbarker.bsky.social and Ashley Nicholls for their incredible work on this project, and to the Jasmin Fisher lab and all our collaborators!
By predicting the effect of combining targeted therapies with radiotherapy, as well as over 10,000 combinations of drugs and driver mutations, we find promising strategies to improve treatment outcomes, and identify a 19-gene signature that could help predict response to radiotherapy.

January 12, 2025 at 5:29 PM
By predicting the effect of combining targeted therapies with radiotherapy, as well as over 10,000 combinations of drugs and driver mutations, we find promising strategies to improve treatment outcomes, and identify a 19-gene signature that could help predict response to radiotherapy.
Huge thanks to everyone who made the work above possible, especially the @cambiochem.bsky.social , @ucllifesciences.bsky.social, Joseph Bloom, Jasmin Fisher and the Fisher Lab (www.ucl.ac.uk/cancer/resea...)

January 12, 2025 at 3:19 PM
Huge thanks to everyone who made the work above possible, especially the @cambiochem.bsky.social , @ucllifesciences.bsky.social, Joseph Bloom, Jasmin Fisher and the Fisher Lab (www.ucl.ac.uk/cancer/resea...)
Applying my experience in biological networks to LLMs, I find a promising method for extracting interpretable features, Sparse AutoEncoders, is more complex than expected. SAE latents are not always independent, forming clusters mapping interpretable subspaces. (www.lesswrong.com/posts/WNoqEi...)

January 12, 2025 at 3:19 PM
Applying my experience in biological networks to LLMs, I find a promising method for extracting interpretable features, Sparse AutoEncoders, is more complex than expected. SAE latents are not always independent, forming clusters mapping interpretable subspaces. (www.lesswrong.com/posts/WNoqEi...)
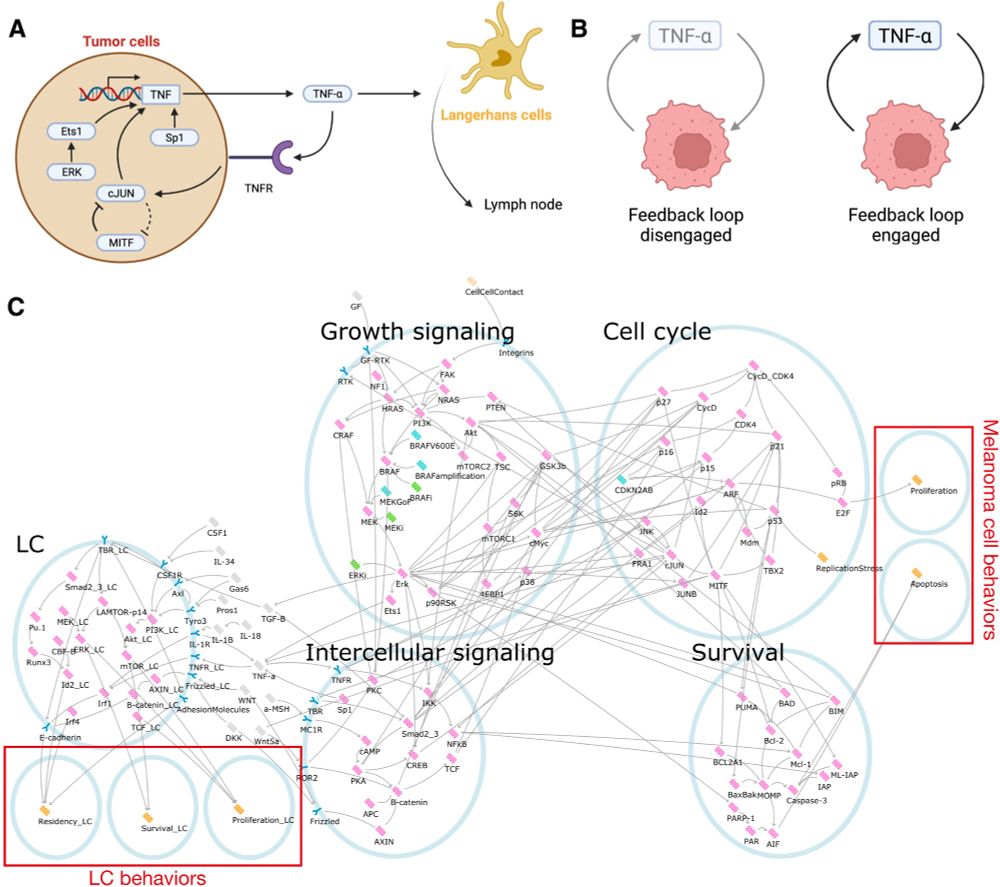
Melanomas often escape immune surveillance. Rowan Howell led work to discover why using network modelling, finding that this rests on a TNF-alpha feedback loop that prevents Langerhans cell migration to the lymph nodes. (www.science.org/doi/full/10....)
January 12, 2025 at 3:19 PM
Melanomas often escape immune surveillance. Rowan Howell led work to discover why using network modelling, finding that this rests on a TNF-alpha feedback loop that prevents Langerhans cell migration to the lymph nodes. (www.science.org/doi/full/10....)
The behaviour of myeloproliferative neoplasms depends not only on what mutations they have, but the order in which they acquired them. Laure Talarmain led research to work out why, finding that HOXA9 acts like a switch that remembers which mutation occurred first. (www.nature.com/articles/s41...)

January 12, 2025 at 3:19 PM
The behaviour of myeloproliferative neoplasms depends not only on what mutations they have, but the order in which they acquired them. Laure Talarmain led research to work out why, finding that HOXA9 acts like a switch that remembers which mutation occurred first. (www.nature.com/articles/s41...)
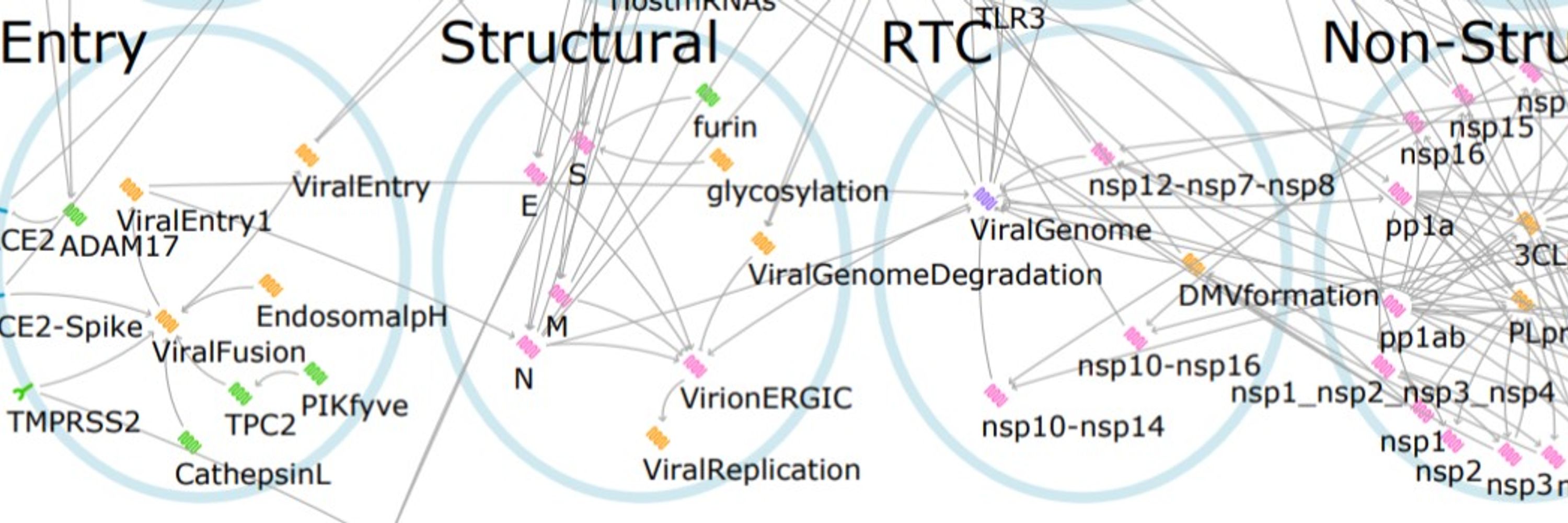
During the pandemic, we rapidly pivoted our methods for finding cancer combination therapy to COVID-19, identifying treatments most likely to be effective for different stages of the disease, as well as remaining useful even with likely resistance mutations. (www.nature.com/articles/s41...)

January 12, 2025 at 3:19 PM
During the pandemic, we rapidly pivoted our methods for finding cancer combination therapy to COVID-19, identifying treatments most likely to be effective for different stages of the disease, as well as remaining useful even with likely resistance mutations. (www.nature.com/articles/s41...)
Professor Jasmin Fisher and I reviewed the exciting progress in using executable computational models to understand cancer complexity. These offer the opportunity to integrate diverse data to predict response and resistance, paving the way for personalized treatment (www.nature.com/articles/s41...)

January 12, 2025 at 3:19 PM
Professor Jasmin Fisher and I reviewed the exciting progress in using executable computational models to understand cancer complexity. These offer the opportunity to integrate diverse data to predict response and resistance, paving the way for personalized treatment (www.nature.com/articles/s41...)
We investigated cooperation between cancer cells in silico and in vivo, showing how high-Myc expressing cells drive tumour growth but become dependent on WNT-expressing cells, and found combinations of drugs that best disrupted this cooperation. (www.pnas.org/doi/abs/10.1...)

January 12, 2025 at 3:19 PM
We investigated cooperation between cancer cells in silico and in vivo, showing how high-Myc expressing cells drive tumour growth but become dependent on WNT-expressing cells, and found combinations of drugs that best disrupted this cooperation. (www.pnas.org/doi/abs/10.1...)
Based on work by Garg et al. (academic.oup.com/bioinformati...), we took advantage of binary decision diagrams to efficiently search future and past possible states of genetic regulatory to understand how past mutations restrict future evolution. (api.repository.cam.ac.uk/server/api/c...)

January 12, 2025 at 3:19 PM
Based on work by Garg et al. (academic.oup.com/bioinformati...), we took advantage of binary decision diagrams to efficiently search future and past possible states of genetic regulatory to understand how past mutations restrict future evolution. (api.repository.cam.ac.uk/server/api/c...)

