Lucas Farnung
@lucas.farnunglab.com
Assistant Professor of Cell Biology at Harvard Medical School | HHMI Freeman Hrabowski Scholar | Nexus of chromatin, transcription, replication, and epigenetics. farnunglab.com
Della Syau successfully defended her PhD thesis today. Congrats, Dr. Syau. Strike #3!

October 9, 2025 at 12:02 AM
Della Syau successfully defended her PhD thesis today. Congrats, Dr. Syau. Strike #3!
New week, new tool: Find our Protein Domain Designer tool to generate publication-ready protein domain diagrams here: domaindesigner.farnunglab.com

September 29, 2025 at 3:01 PM
New week, new tool: Find our Protein Domain Designer tool to generate publication-ready protein domain diagrams here: domaindesigner.farnunglab.com
🚀 Ordering lab supplies just got way easier in our lab.
We built a workflow (github.com/farnunglab/N...) where tapping an NFC tag on a reagent bottle automatically triggers an order request in Quartzy. No more scribbling notes or hunting for a computer. Here’s how 👇
We built a workflow (github.com/farnunglab/N...) where tapping an NFC tag on a reagent bottle automatically triggers an order request in Quartzy. No more scribbling notes or hunting for a computer. Here’s how 👇

September 26, 2025 at 1:15 PM
🚀 Ordering lab supplies just got way easier in our lab.
We built a workflow (github.com/farnunglab/N...) where tapping an NFC tag on a reagent bottle automatically triggers an order request in Quartzy. No more scribbling notes or hunting for a computer. Here’s how 👇
We built a workflow (github.com/farnunglab/N...) where tapping an NFC tag on a reagent bottle automatically triggers an order request in Quartzy. No more scribbling notes or hunting for a computer. Here’s how 👇
Martin Filipovski successfully defended his PhD thesis today! Congrats, Dr. Filipovski.


September 4, 2025 at 9:55 PM
Martin Filipovski successfully defended his PhD thesis today! Congrats, Dr. Filipovski.
In our IWS1 story, we also visualize the histone reader LEDGF PWWP domain on a transcribed downstream nucleosome. Because the PWWP requires both DNA gyres for binding, it binds to the promoter-distal side of the nucleosome during transcription through the nucleosome.

September 3, 2025 at 3:42 PM
In our IWS1 story, we also visualize the histone reader LEDGF PWWP domain on a transcribed downstream nucleosome. Because the PWWP requires both DNA gyres for binding, it binds to the promoter-distal side of the nucleosome during transcription through the nucleosome.
🧬 Transcription elongation by RNA polymerase II relies on a web of elongation factors. Our new work shows how IWS1 acts as a modular scaffold to stabilize & stimulate elongation. Fantastic work by Della Syau! www.biorxiv.org/content/10.1...
August 29, 2025 at 10:14 AM
🧬 Transcription elongation by RNA polymerase II relies on a web of elongation factors. Our new work shows how IWS1 acts as a modular scaffold to stabilize & stimulate elongation. Fantastic work by Della Syau! www.biorxiv.org/content/10.1...
Congratulations to our own @sophiero.bsky.social and team for achieving 3rd place 🥉 in the Merck Innovation Cup 2025. The entire lab is very proud of Sophie's achievement!

July 25, 2025 at 6:00 PM
Congratulations to our own @sophiero.bsky.social and team for achieving 3rd place 🥉 in the Merck Innovation Cup 2025. The entire lab is very proud of Sophie's achievement!
First PhD candidate (congrats, Dr. James!) to graduate today and check out this beautiful nucleosome/ocean cake made by PhD candidate Della Syau:

June 16, 2025 at 9:06 PM
First PhD candidate (congrats, Dr. James!) to graduate today and check out this beautiful nucleosome/ocean cake made by PhD candidate Della Syau:
Congratulations to 🎓 Dr. Allison James 🎉. Allison is the first PhD candidate to graduate from the Farnung Lab!


June 16, 2025 at 6:39 PM
Congratulations to 🎓 Dr. Allison James 🎉. Allison is the first PhD candidate to graduate from the Farnung Lab!

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
May 6, 2025 at 3:08 PM

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
April 6, 2025 at 9:45 AM
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
🚨New paper🚨Our new study with the lab of Phil Cole uncovers how SIRT6 dynamically adapts structurally and enzymatically to target diverse histone acylations (e.g., H3K27ac) within nucleosomes. @jonmarkert.bsky.social
doi.org/10.1016/j.jb... #Chromatin #SIRT6 #CryoEM
doi.org/10.1016/j.jb... #Chromatin #SIRT6 #CryoEM
March 28, 2025 at 3:44 PM
🚨New paper🚨Our new study with the lab of Phil Cole uncovers how SIRT6 dynamically adapts structurally and enzymatically to target diverse histone acylations (e.g., H3K27ac) within nucleosomes. @jonmarkert.bsky.social
doi.org/10.1016/j.jb... #Chromatin #SIRT6 #CryoEM
doi.org/10.1016/j.jb... #Chromatin #SIRT6 #CryoEM
Another highlight from our story: @jonmarkert.bsky.social uncovered how SPT6 binds the exposed H2A–H2B dimer during nucleosomal traversal by Pol II. AlphaFold helped tremendously in validating this interaction. tinyurl.com/setd2 Also check out our H2A–H2B screen: www.biorxiv.org/content/10.1...

December 18, 2024 at 2:08 PM
Another highlight from our story: @jonmarkert.bsky.social uncovered how SPT6 binds the exposed H2A–H2B dimer during nucleosomal traversal by Pol II. AlphaFold helped tremendously in validating this interaction. tinyurl.com/setd2 Also check out our H2A–H2B screen: www.biorxiv.org/content/10.1...
In our SETD2 paper, we visualize a novel nucleosome transfer intermediate during txn. It is the earliest intermediate visualized yet. In the intermediate, the histone chaperone FACT coordinates histone binding with a previously uncharacterized RTF1 helix. All details in the paper: tinyurl.com/setd2

December 12, 2024 at 7:30 PM
In our SETD2 paper, we visualize a novel nucleosome transfer intermediate during txn. It is the earliest intermediate visualized yet. In the intermediate, the histone chaperone FACT coordinates histone binding with a previously uncharacterized RTF1 helix. All details in the paper: tinyurl.com/setd2
Ever wondered how transcription choreographs histone modifications? Our work reveals the basis of co-transcriptional H3K36me3 by SETD2. We visualize how a histone writer coordinates with the transcription machinery! This is the magnus opus of @jonmarkert.bsky.social!
tinyurl.com/setd2
tinyurl.com/setd2
December 12, 2024 at 7:16 PM
Ever wondered how transcription choreographs histone modifications? Our work reveals the basis of co-transcriptional H3K36me3 by SETD2. We visualize how a histone writer coordinates with the transcription machinery! This is the magnus opus of @jonmarkert.bsky.social!
tinyurl.com/setd2
tinyurl.com/setd2
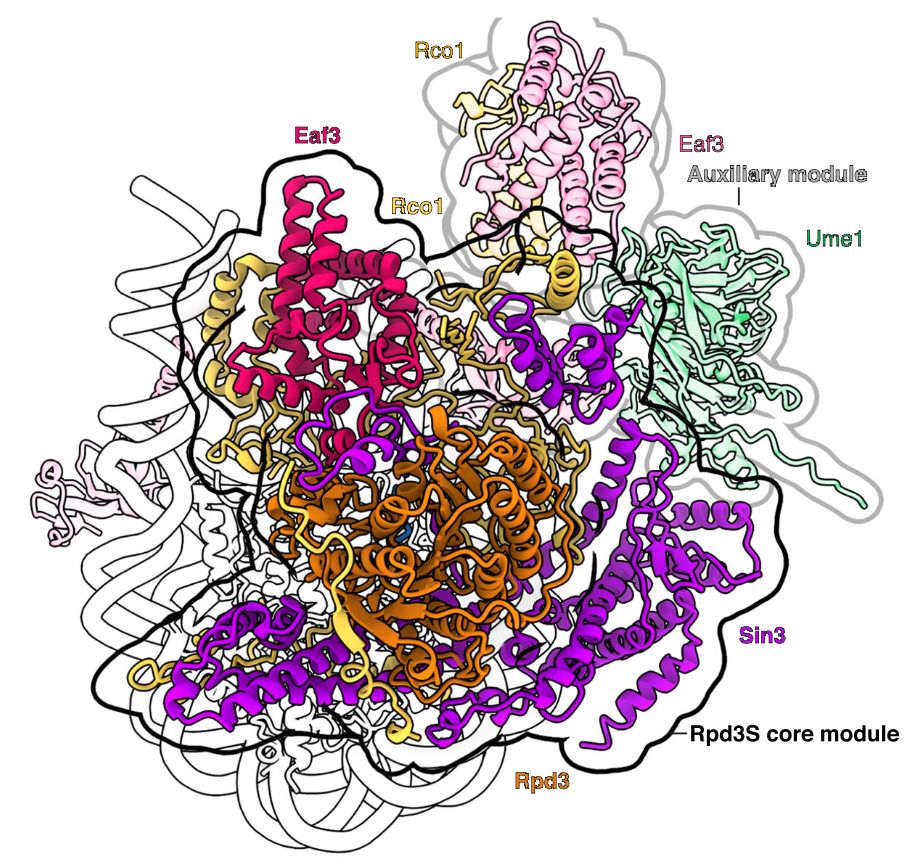
Our paper on the structure of the complete Rpd3S-nucleosome complex is now out in Nat Comm: rdcu.be/ds6k1 . Congrats to @jonmarkert.bsky.social
and co-corresponding author Seychelle Vos! This is the second HDAC-nucleosome structure from Jon this year. Amazing!
and co-corresponding author Seychelle Vos! This is the second HDAC-nucleosome structure from Jon this year. Amazing!
December 8, 2023 at 12:32 PM
Our paper on the structure of the complete Rpd3S-nucleosome complex is now out in Nat Comm: rdcu.be/ds6k1 . Congrats to @jonmarkert.bsky.social
and co-corresponding author Seychelle Vos! This is the second HDAC-nucleosome structure from Jon this year. Amazing!
and co-corresponding author Seychelle Vos! This is the second HDAC-nucleosome structure from Jon this year. Amazing!
Doing much better on antibodies:

October 31, 2023 at 1:35 PM
Doing much better on antibodies:

