
We also find data to support our model: we performed a systematic literature search, which shows that generalists (producing both enzymes) dominate among chitin-degrading microbes. 5/6

We also find data to support our model: we performed a systematic literature search, which shows that generalists (producing both enzymes) dominate among chitin-degrading microbes. 5/6
Our analysis paints a fascinating picture: interactions between endo- and exo-enzyme producers can flip from positive to negative depending on substrate levels! 4/6
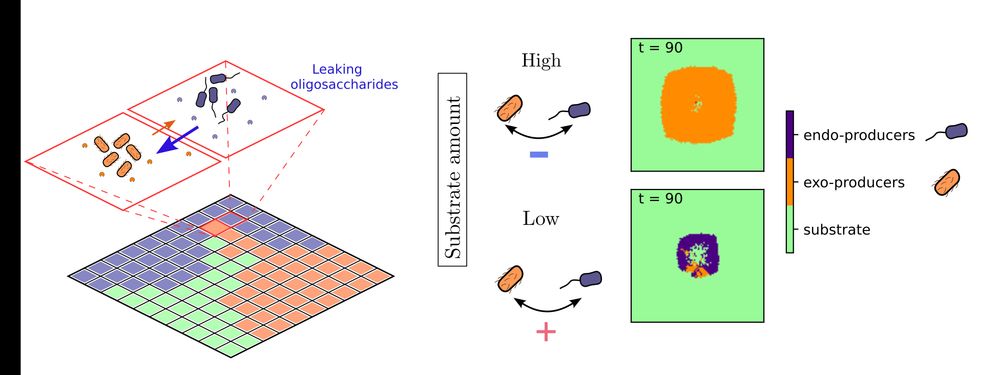
Our analysis paints a fascinating picture: interactions between endo- and exo-enzyme producers can flip from positive to negative depending on substrate levels! 4/6
1. Of exo-enzymes thrive when many polymer chains are around.
2. Of endo-enzymes unexpectedly excel only when fewer chains are present.
3. Of both enzymes together expand their niche and may benefit from enzyme synergy. 3/6

1. Of exo-enzymes thrive when many polymer chains are around.
2. Of endo-enzymes unexpectedly excel only when fewer chains are present.
3. Of both enzymes together expand their niche and may benefit from enzyme synergy. 3/6
1. “Exo”-enzymes snip small units from the ends of polymer chains.
2. “Endo”-enzymes break polymers at random, creating varied-sized fragments.
Using a theoretical model, we simulated polymer degradation and microbial growth. 2/6
1. “Exo”-enzymes snip small units from the ends of polymer chains.
2. “Endo”-enzymes break polymers at random, creating varied-sized fragments.
Using a theoretical model, we simulated polymer degradation and microbial growth. 2/6

