Kevin Drew
@ksdrew.bsky.social
Assistant Professor at University of Illinois at Chicago (UIC) in Biological Sciences focused on macromolecular assemblies.
Finally, we utilize our network to build structural models of a ciliary protein complex linked to oral-facial-digital syndrome. We identified several human pathogenic mutations at the interface of OFD1 and FOPNL (CEP20), pointing to a mechanism of pathology.

July 29, 2025 at 8:58 PM
Finally, we utilize our network to build structural models of a ciliary protein complex linked to oral-facial-digital syndrome. We identified several human pathogenic mutations at the interface of OFD1 and FOPNL (CEP20), pointing to a mechanism of pathology.
Encouraged by this, we prioritized ~2,500 high-confidence DirectContacts2 protein pairs and ran them through AF3. We see excellent enrichment of high quality models consistent with the AF2 models.

July 29, 2025 at 8:58 PM
Encouraged by this, we prioritized ~2,500 high-confidence DirectContacts2 protein pairs and ran them through AF3. We see excellent enrichment of high quality models consistent with the AF2 models.
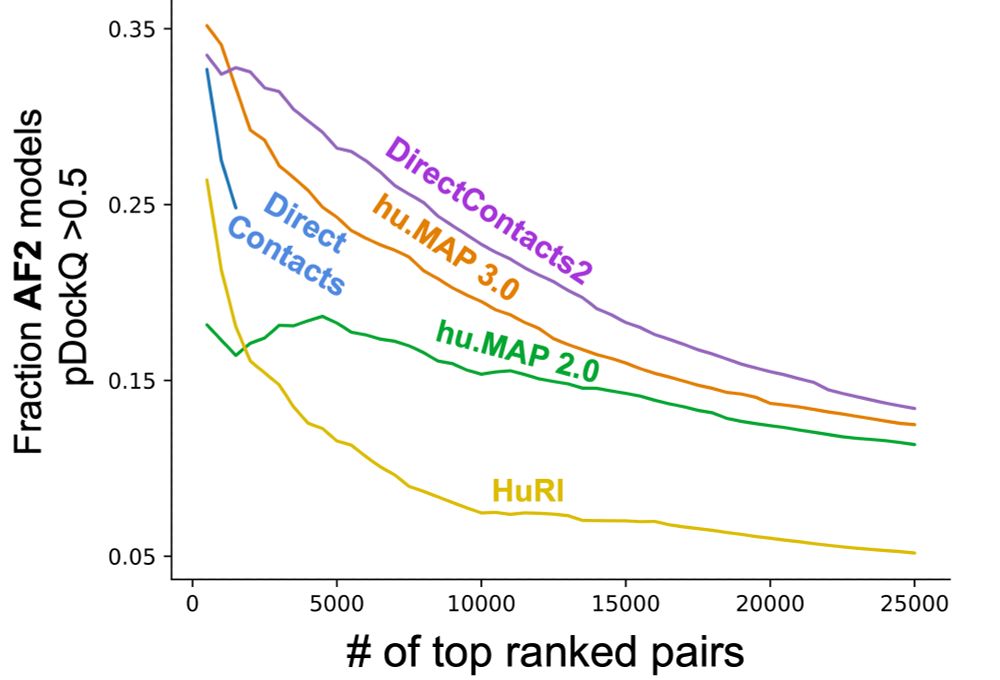
First, using a compendium of computed structural models for evaluation, we see our DirectContacts2 network outperforms other networks at prioritizing protein pairs for AlphaFold2 modeling.
July 29, 2025 at 8:58 PM
First, using a compendium of computed structural models for evaluation, we see our DirectContacts2 network outperforms other networks at prioritizing protein pairs for AlphaFold2 modeling.
We evaluated our network on a benchmark of known direct interactions from the PDB and see that it outperforms other networks including our previous attempt at solving this problem (DirectContacts doi.org/10.1371/jour...) and our hu.MAP co-complex networks!

July 29, 2025 at 8:58 PM
We evaluated our network on a benchmark of known direct interactions from the PDB and see that it outperforms other networks including our previous attempt at solving this problem (DirectContacts doi.org/10.1371/jour...) and our hu.MAP co-complex networks!
Determining whether two proteins have a direct physical interaction is an important (and difficult!) problem as protein complex assembly relies on physical interactions and protein interaction interfaces are the target of many therapeutics.

July 29, 2025 at 8:58 PM
Determining whether two proteins have a direct physical interaction is an important (and difficult!) problem as protein complex assembly relies on physical interactions and protein interaction interfaces are the target of many therapeutics.
We see SEPTIN6 is expressed in blood lineages but not detected in lineages with SEPTIN10 such as gastric and esophageal cells. This analysis provides further support of mutually exclusive proteins and points to specific lineages where a complex’s function may be altered.

May 29, 2025 at 11:24 PM
We see SEPTIN6 is expressed in blood lineages but not detected in lineages with SEPTIN10 such as gastric and esophageal cells. This analysis provides further support of mutually exclusive proteins and points to specific lineages where a complex’s function may be altered.
Mutually exclusive proteins introduce a conflict for the organism as they cannot both bind the common subunit at the same time. We find this conflict is resolved primarily by differences in expression of the subunits.

May 29, 2025 at 11:24 PM
Mutually exclusive proteins introduce a conflict for the organism as they cannot both bind the common subunit at the same time. We find this conflict is resolved primarily by differences in expression of the subunits.
Using AlphaFold models from @bioinfo.se
and @pedrobeltrao.bsky.social’s work, we identified 5,800+ mutually exclusive protein interactions where two proteins have overlapping interfaces with another protein. This was greatly expanded during review using Jänes et al's resource (PMC11160786).
and @pedrobeltrao.bsky.social’s work, we identified 5,800+ mutually exclusive protein interactions where two proteins have overlapping interfaces with another protein. This was greatly expanded during review using Jänes et al's resource (PMC11160786).

May 29, 2025 at 11:24 PM
Using AlphaFold models from @bioinfo.se
and @pedrobeltrao.bsky.social’s work, we identified 5,800+ mutually exclusive protein interactions where two proteins have overlapping interfaces with another protein. This was greatly expanded during review using Jänes et al's resource (PMC11160786).
and @pedrobeltrao.bsky.social’s work, we identified 5,800+ mutually exclusive protein interactions where two proteins have overlapping interfaces with another protein. This was greatly expanded during review using Jänes et al's resource (PMC11160786).
For example, we identify the uncharacterized protein TMEM167A as co-complex with the disease gene, IER3IP1 (i.e. MED Syndrome). Using AlphaFold, we modeled the interaction of these two proteins and observe patient mutations at the interaction interface.

May 29, 2025 at 11:24 PM
For example, we identify the uncharacterized protein TMEM167A as co-complex with the disease gene, IER3IP1 (i.e. MED Syndrome). Using AlphaFold, we modeled the interaction of these two proteins and observe patient mutations at the interaction interface.
For example, we see ATP synthase and NADH dehydrogenase covary together as they are both involved in OXPHOS. We also see the proteasome, translational machinery and, interestingly, the MCM complex covary together.

May 29, 2025 at 11:24 PM
For example, we see ATP synthase and NADH dehydrogenase covary together as they are both involved in OXPHOS. We also see the proteasome, translational machinery and, interestingly, the MCM complex covary together.
hu.MAP3.0 is substantially updated from hu.MAP2.0 in terms of total experiments (adding and reanalyzing 10k experiments from BioPlex (@edhuttlin.bsky.social) as well as using an improved ML workflow based on Autogluon. This improves on both precision and recall.

May 29, 2025 at 11:24 PM
hu.MAP3.0 is substantially updated from hu.MAP2.0 in terms of total experiments (adding and reanalyzing 10k experiments from BioPlex (@edhuttlin.bsky.social) as well as using an improved ML workflow based on Autogluon. This improves on both precision and recall.
Thank you for organizing Anna!

March 7, 2025 at 8:45 PM
Thank you for organizing Anna!


