
PhD @CEITEC.eu @masarykuniversity.bsky.social
About me:
https://sites.google.com/view/krishnendubera

www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
In collaboration with Gregor Anderlu, NIC, Slo read 👇 www.science.org/doi/10.1126/...
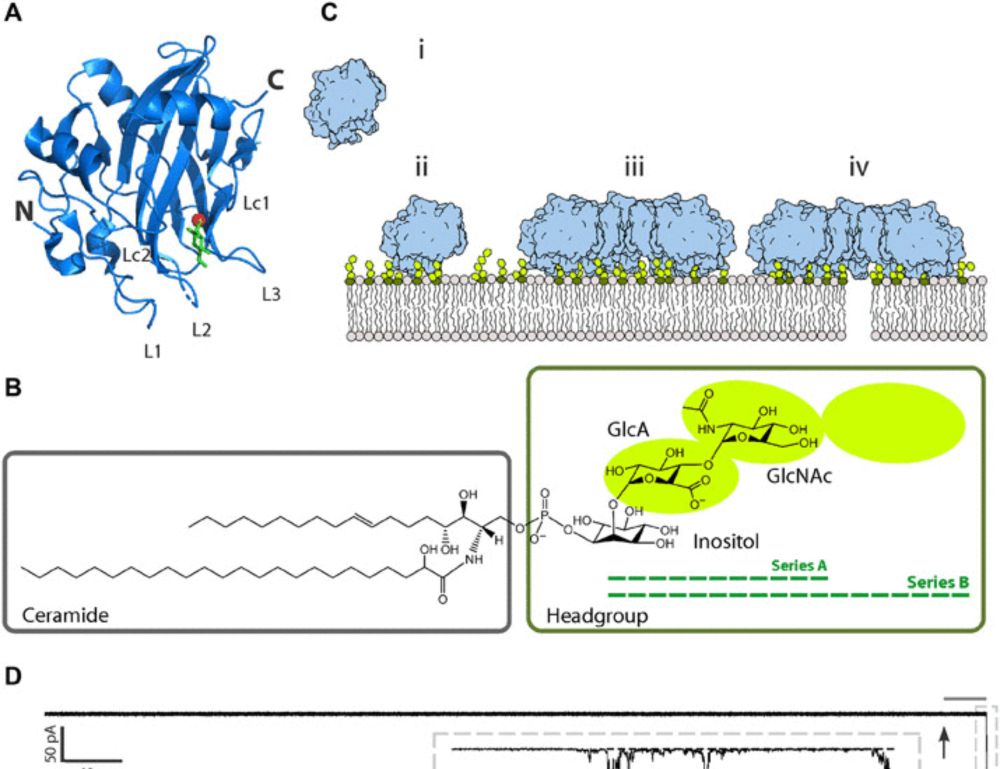
In collaboration with Gregor Anderlu, NIC, Slo read 👇 www.science.org/doi/10.1126/...





Thanks all co-authors. Thanks #CzechELib for open access support!
Link: shorturl.at/DhpAz
#AgScience #InsecticideDesign #ACS #JAgFoodChem #CEITEC_Brno #muni_cz #CzechAcademy #MasarykUni
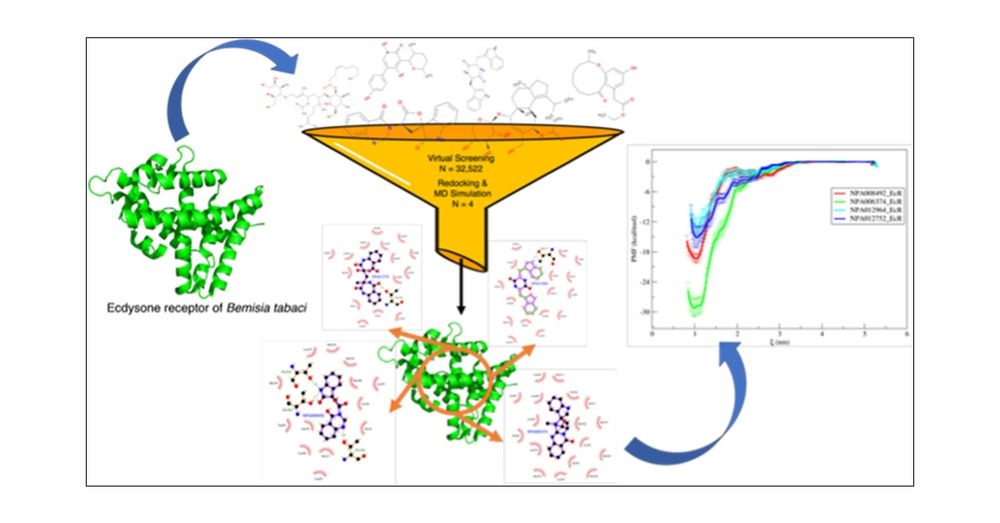
Thanks all co-authors. Thanks #CzechELib for open access support!
Link: shorturl.at/DhpAz
#AgScience #InsecticideDesign #ACS #JAgFoodChem #CEITEC_Brno #muni_cz #CzechAcademy #MasarykUni
@angelaparis3.bsky.social , in collaboration with University of Trieste, University of Udine, CNR-IC
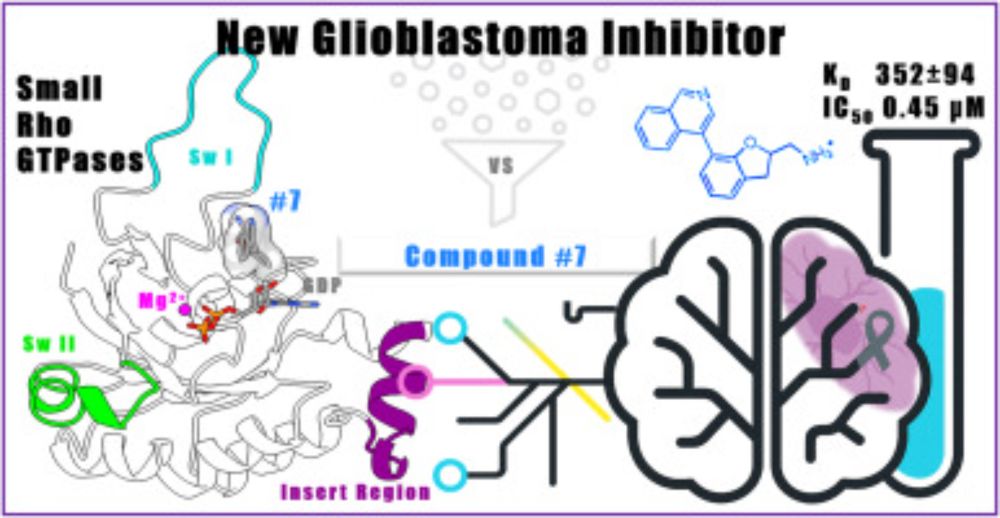
@angelaparis3.bsky.social , in collaboration with University of Trieste, University of Udine, CNR-IC
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
📖 Read it here: rdcu.be/ef6YX
📝 Support the statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #collaboration
📖 Read it here: rdcu.be/ef6YX
📝 Support the statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #collaboration
@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...

@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...
#RNA #MembraneBiology #compchem #compbiol
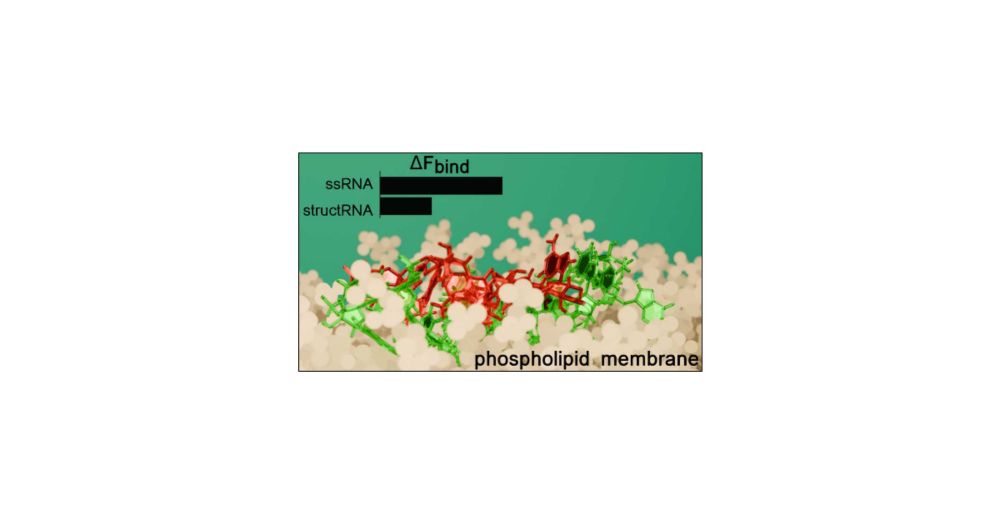
#RNA #MembraneBiology #compchem #compbiol
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs

Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
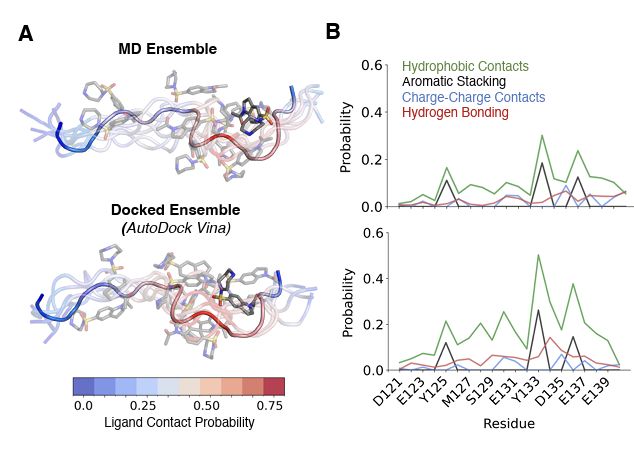
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
www.euchems.eu/divisions/ch...

www.euchems.eu/divisions/ch...
pubs.acs.org/doi/10.1021/...
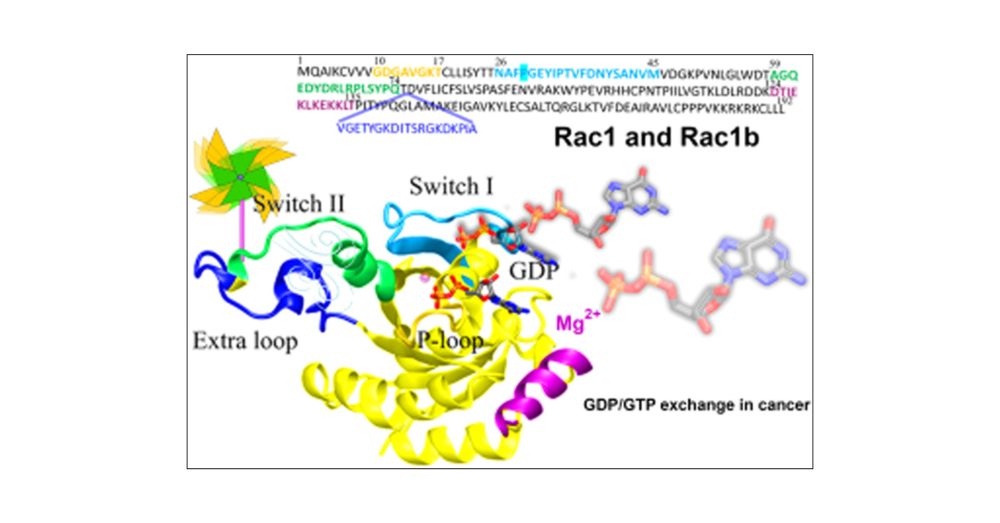
pubs.acs.org/doi/10.1021/...

We have studied Polyethylene Glycol‐Based Refolding Kinetic Modulation of CRABP I Protein in this article. Details 👇👇
analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/...
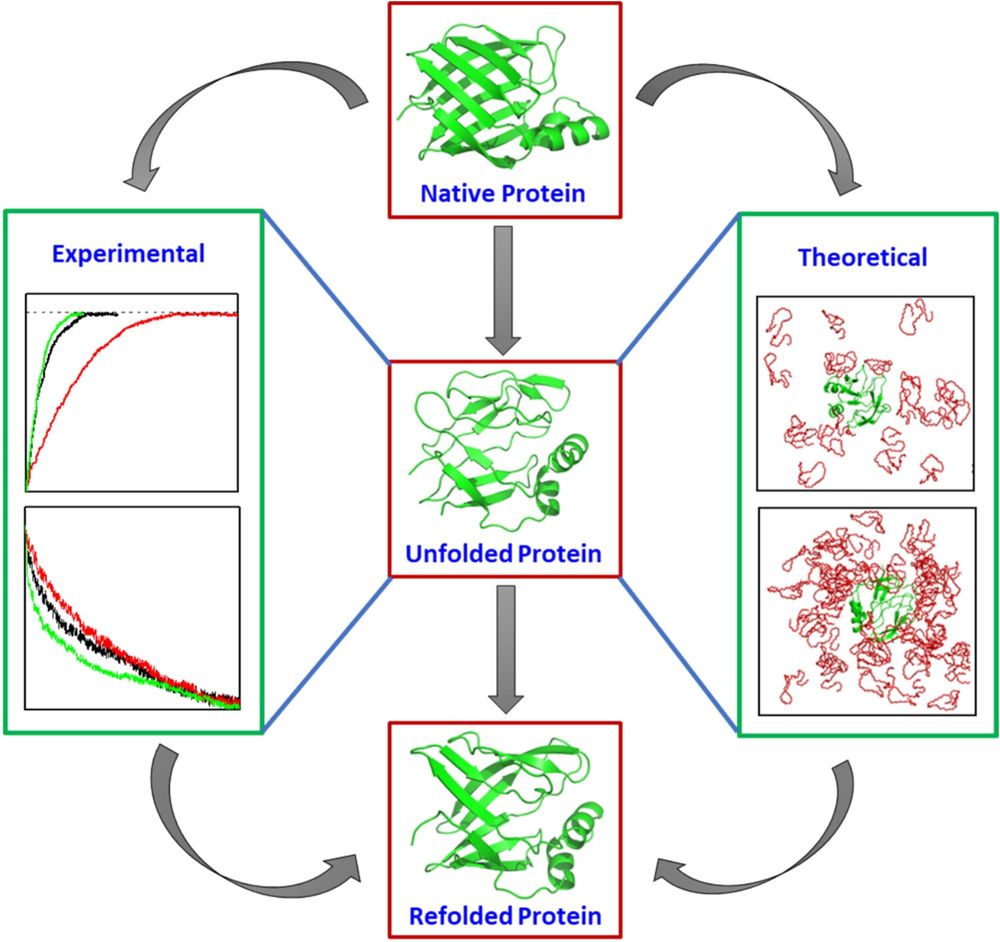
We have studied Polyethylene Glycol‐Based Refolding Kinetic Modulation of CRABP I Protein in this article. Details 👇👇
analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/...
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...


