
https://orcid.org/0000-0001-5470-8299
WGS data: pacificbiosciences.github.io/ParaviewerWG...
PureTarget data: pacificbiosciences.github.io/ParaviewerPT...
WGS data: pacificbiosciences.github.io/ParaviewerWG...
PureTarget data: pacificbiosciences.github.io/ParaviewerPT...


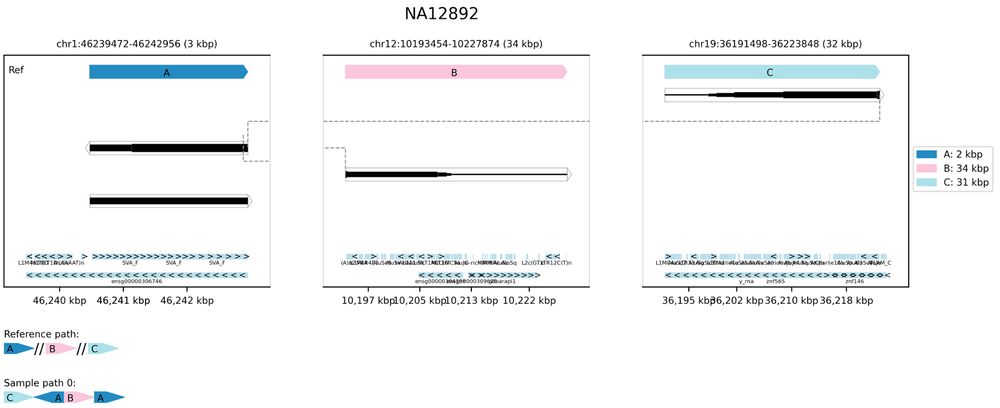


If you were interested but missed it: www.pacb.com/wp-content/u...
If you were interested but missed it: www.pacb.com/wp-content/u...

