Trying to understand and engineer our wonderfully weird genomes 🧬
🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇

🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇
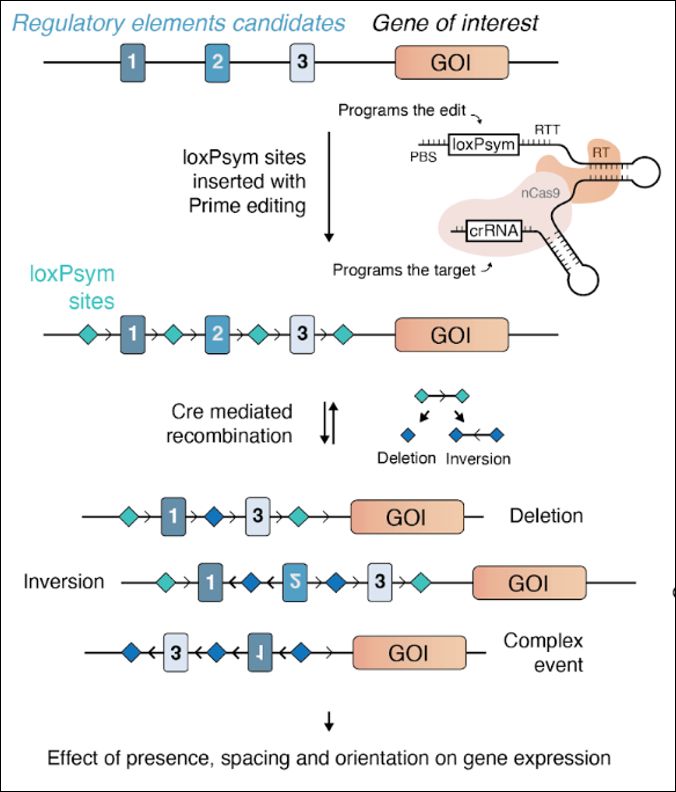
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
lnkd.in/da-gitNc
lnkd.in/da-gitNc
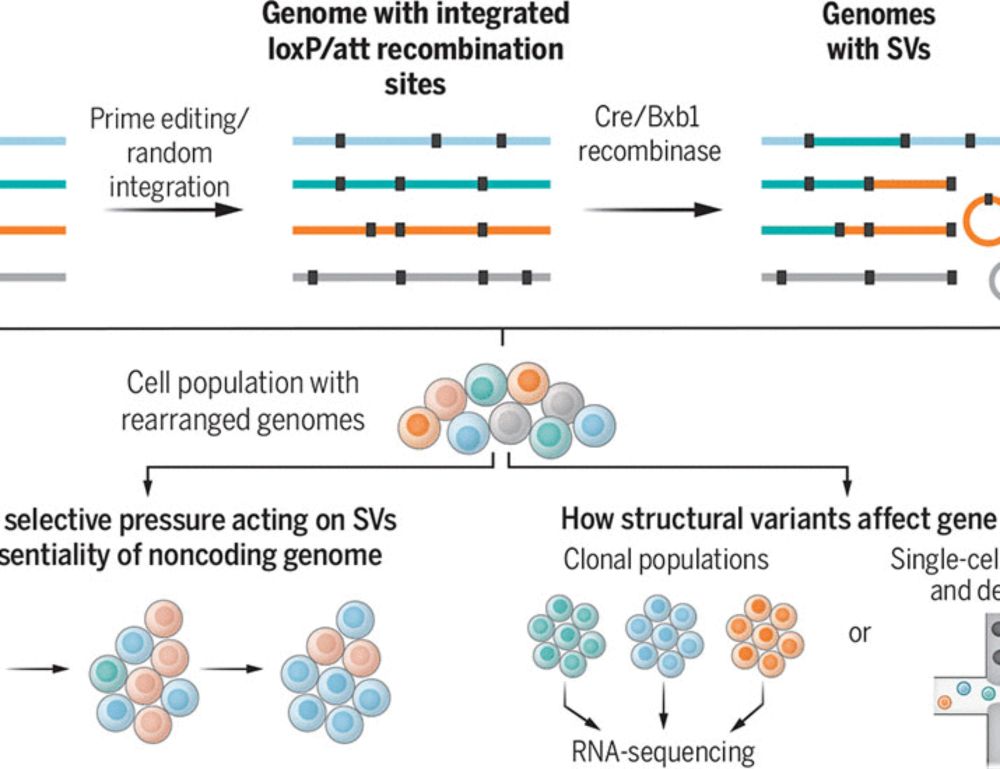
www.science.org/doi/10.1126/...
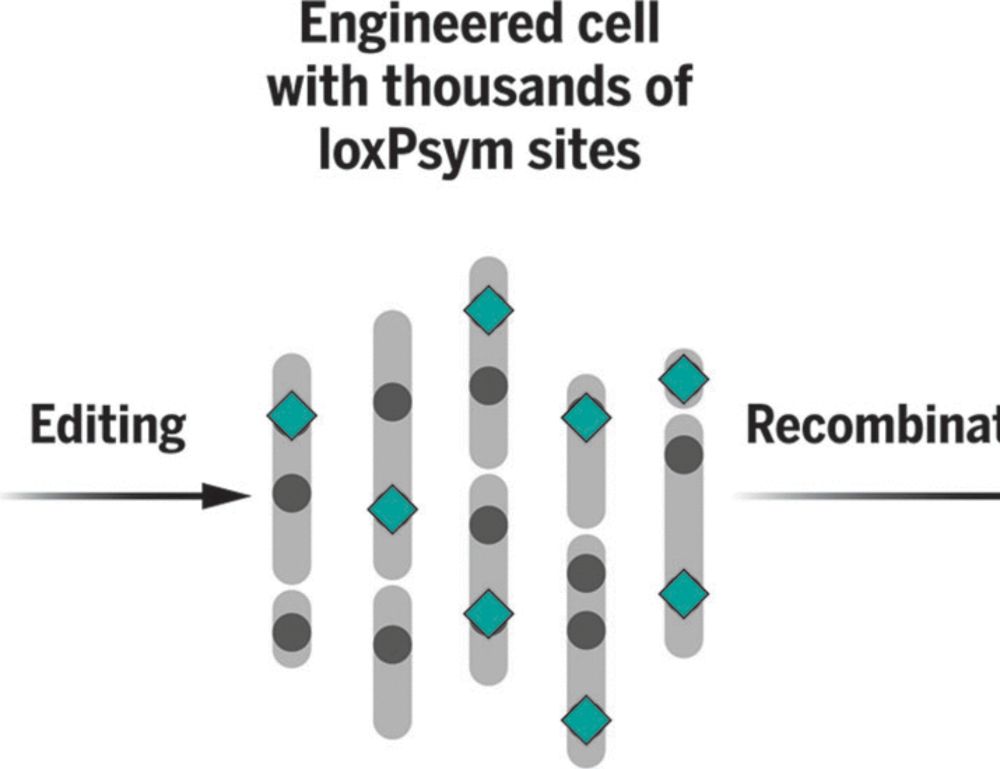
www.pinglay-lab.com
www.pinglay-lab.com
www.science.org/doi/10.1126/...
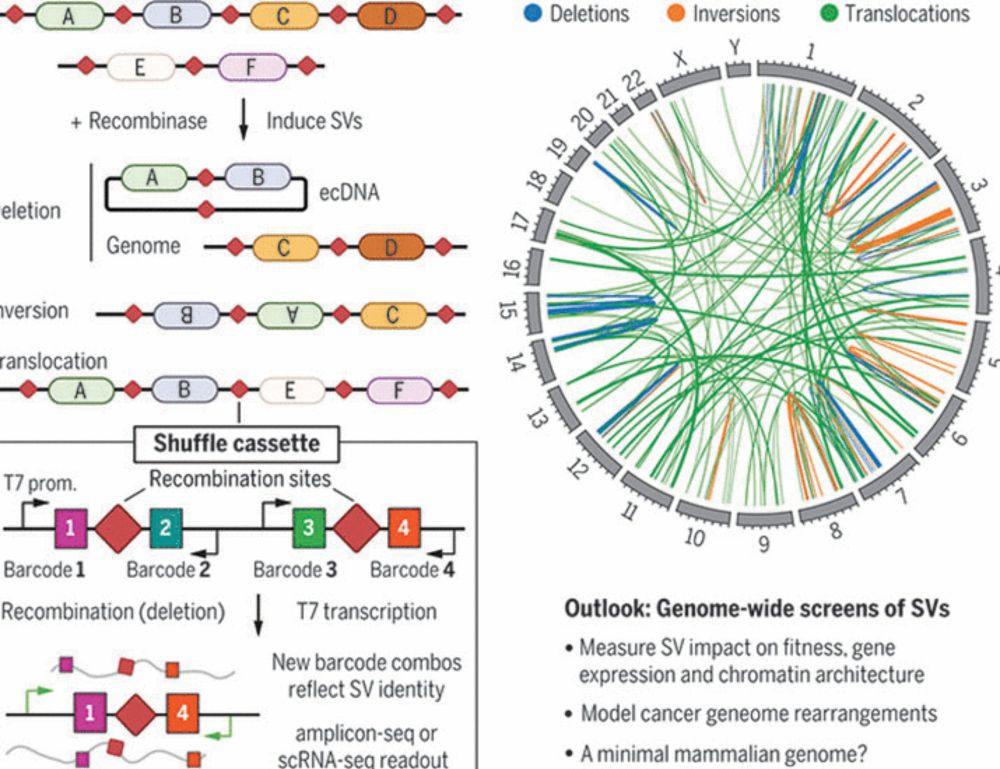
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/... and www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/... and www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
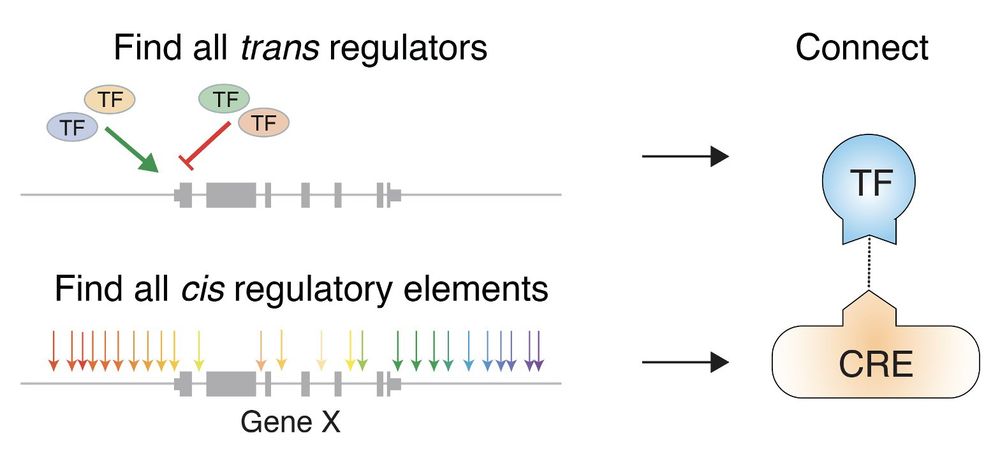
In this work, superstar postdoc @xinhexue.bsky.social combined 2 kinds of pooled CRISPR screens to pinpoint noncoding regulatory elements and the transcription factors that activate these elements.