Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
690 followers
3.3K following
130 posts
Doing a Ph.D. AI in Bio. | Ex @WhiteLabGx @BroadInstitute @MIT | Built @PiPleteam | ML, Cancer, Genomics, Data Sci, Entrepreneur, FullStack Dev | All views are mine
Posts
Media
Videos
Starter Packs
Pinned
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Nov 19
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Oct 6
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Oct 6
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Oct 6
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jul 24
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jul 24
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jul 24
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jul 21
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jul 17
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jun 20
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jun 20
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jun 20
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jun 17
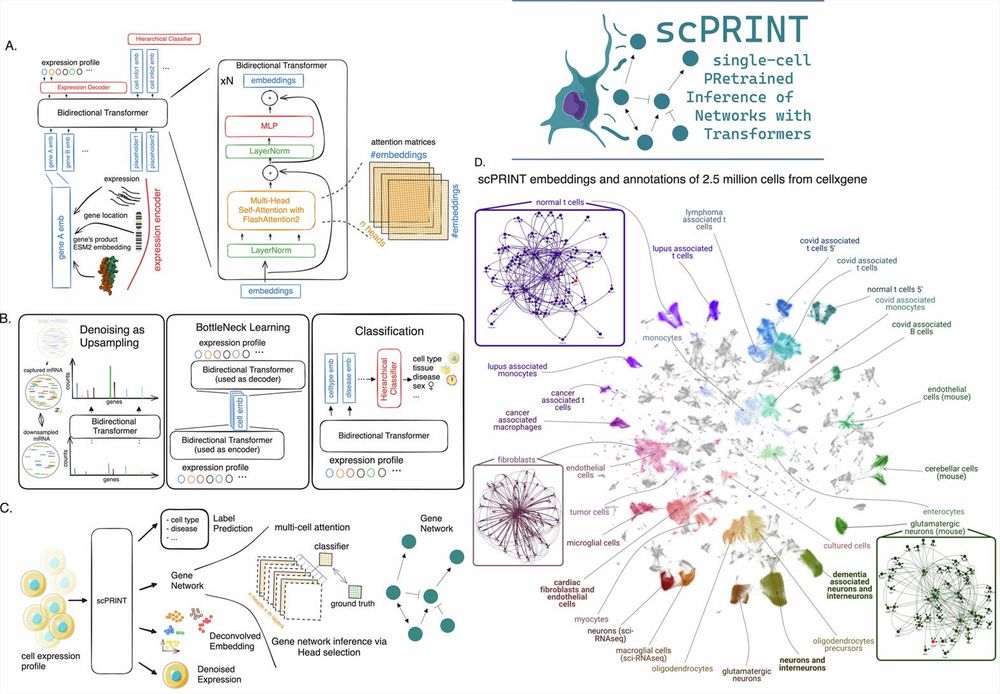
scPRINT: pre-training on 50 million cells allows robust gene network predictions
Nature Communications - Authors present a state-of-the-art cell foundation model trained on 50 million cells. They show that the model generates a meaningful gene network and has zero-shot...
www.nature.com
Jeremie Kalfon 👨💻🧬🤖🚀
@jkobject.com
· Jun 17

scPRINT | v1.0 | Virtual Cells Platform
scPRINT is a cell foundation model, also called a Large Cell Model (LCM), trained on single-cell RNA sequence (scRNAseq) data from more than 50M human and mouse cells available through CZ CELLxGENE. Based on the transformer architecture, the model is fully open source and reproducible, with multiple checkpoint sizes available from 2M to 100M parameters. scPRINT demonstrated high performance for genome-wide cell-specific gene network inference when benchmarked against state-of-the-art models (e.g., scGPT, Geneformer v2, GENIE3). In addition, scPRINT has various zero-shot capabilities, including cell embedding, cell label prediction (e.g., cell type, sex, disease), and gene expression imputation, highlighting its potential as a versatile tool for single-cell analysis.
virtualcellmodels.cziscience.com