https://scholar.google.com/citations?hl=en&user=PRTcXNsAAAAJ
journals.plos.org/plospathogen...

journals.plos.org/plospathogen...
Congrats to Barbara, Vanisha, Liz, and the rest of the team.
www.nature.com/articles/s41...

Congrats to Barbara, Vanisha, Liz, and the rest of the team.
www.nature.com/articles/s41...
Guided by @courbongautier.bsky.social's structure of myco ATP synthase bound to a squaramide, they have made SQAs that are less toxic, more potent, more stable than AstraZeneca's SQ31f
#MedChem #TB #NTM
pubs.acs.org/doi/full/10....

Guided by @courbongautier.bsky.social's structure of myco ATP synthase bound to a squaramide, they have made SQAs that are less toxic, more potent, more stable than AstraZeneca's SQ31f
#MedChem #TB #NTM
pubs.acs.org/doi/full/10....
Develop and deliver high-impact scientific marketing content, illustrate the value of new #CryoSPARC features and #cryoEM advances, and help scientists understand how our tools can help solve their problems.
structura.bio/careers/appl...

Develop and deliver high-impact scientific marketing content, illustrate the value of new #CryoSPARC features and #cryoEM advances, and help scientists understand how our tools can help solve their problems.
structura.bio/careers/appl...
A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated C. difficile infection in mice.
#MicroSky 🦠💊
www.nature.com/articles/s41...

A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated C. difficile infection in mice.
#MicroSky 🦠💊
www.nature.com/articles/s41...
This ~ 600 kDa enzyme is asymmetric, rotatory, has unusual stoichiometry, and unique E5 subunits.
www.biorxiv.org/content/10.1...
This ~ 600 kDa enzyme is asymmetric, rotatory, has unusual stoichiometry, and unique E5 subunits.
www.biorxiv.org/content/10.1...
On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg

On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg
Discover practical tips and tricks in our new case study using #CryoSPARC v4.7.1 where we find and refine a low-population interaction partner!
guide.cryosparc.com/processing-d...

Discover practical tips and tricks in our new case study using #CryoSPARC v4.7.1 where we find and refine a low-population interaction partner!
guide.cryosparc.com/processing-d...
@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz

@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz

A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria

A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria
www.biorxiv.org/content/10.1...
#teamtomo

www.biorxiv.org/content/10.1...
#teamtomo
We present guidelines, limitations & future perspectives for processing amyloids in cryoSPARC @structurabio.bsky.social.
Great collaboration of the Kelly Lab and @landerlab.bsky.social #cryoEM #amyloid www.biorxiv.org/content/10.1...

We present guidelines, limitations & future perspectives for processing amyloids in cryoSPARC @structurabio.bsky.social.
Great collaboration of the Kelly Lab and @landerlab.bsky.social #cryoEM #amyloid www.biorxiv.org/content/10.1...

doi.org/10.1101/2025.09.24.678028 #proteindesign

doi.org/10.1101/2025.09.24.678028 #proteindesign
We uncovered step-wise mTORC1 activation on membrane: RAG–Ragulator recruits, RHEB pushes closer, and direct membrane contacts by RAPTOR + mTOR trigger full kinase activity.👉 The membrane is not just a platform — it’s part of the mechanism.
www.nature.com/articles/s41...

We uncovered step-wise mTORC1 activation on membrane: RAG–Ragulator recruits, RHEB pushes closer, and direct membrane contacts by RAPTOR + mTOR trigger full kinase activity.👉 The membrane is not just a platform — it’s part of the mechanism.
www.nature.com/articles/s41...
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
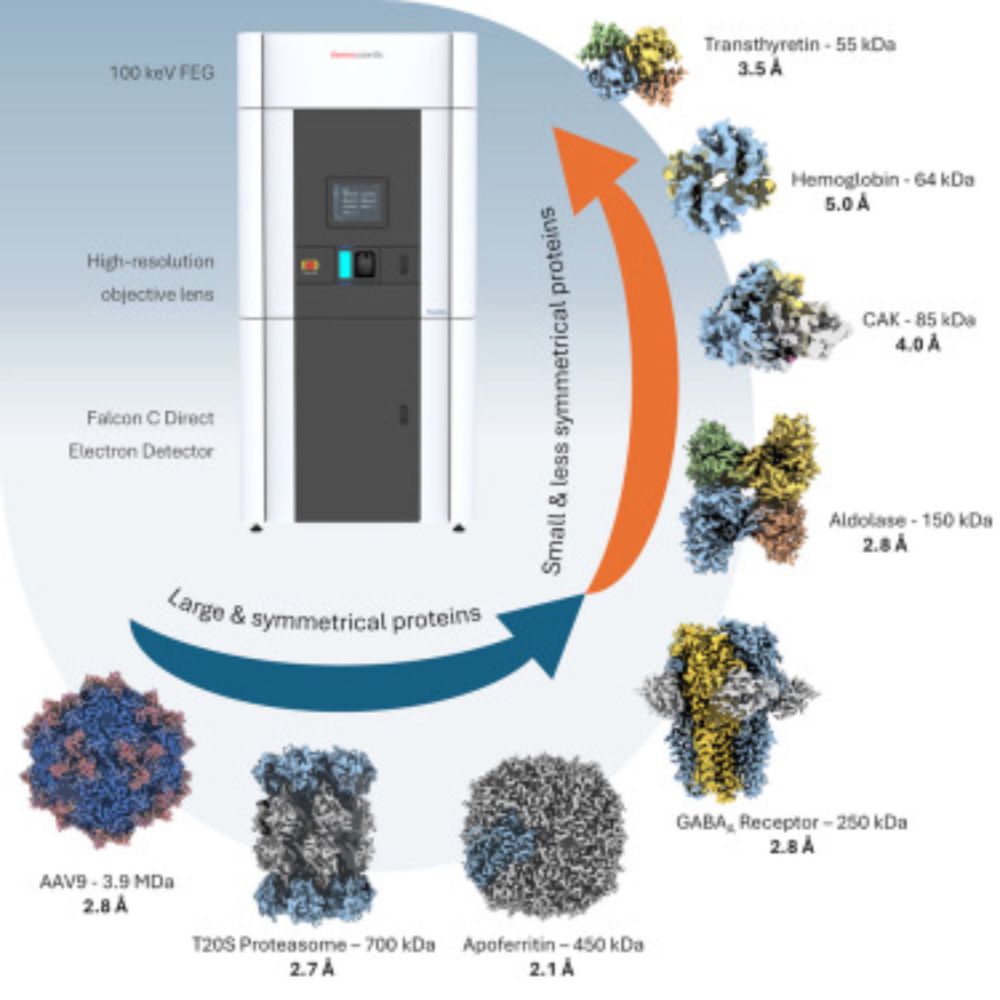
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
go.nature.com/3V8r3Lw

go.nature.com/3V8r3Lw
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo

A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
youtu.be/qQihl6If9vU

youtu.be/qQihl6If9vU
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
guide.cryosparc.com/processing-d...
guide.cryosparc.com/processing-d...
reactor-farnunglab.pythonanywhere.com 1/n
reactor-farnunglab.pythonanywhere.com 1/n

