Jan Felix
@janfelix.bsky.social
Structural Biologist from Gent. Former Post-Doc @ Gutsche group, IBS, Grenoble, FR and @ Savvides Lab, IRC, Gent, BE.
Currently part of the VIB Bioimaging Core Gent Team.
Interested in ❄️🔬 #CryoEM and #Cytokines.
Currently part of the VIB Bioimaging Core Gent Team.
Interested in ❄️🔬 #CryoEM and #Cytokines.
Pinned
Jan Felix
@janfelix.bsky.social
· Sep 2

Now published @cp-cellreports.bsky.social! Antibody-mediated TGF-β1 activation for the treatment of diseases caused by deleterious T cell activity: www.cell.com/cell-reports...
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Reposted by Jan Felix
📢 New preprint:
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇

October 18, 2025 at 6:59 PM
📢 New preprint:
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
Reposted by Jan Felix
Initial attempt at replicating in relion (parameters in next post). This is for Aca2-RNA, using a 100k subset of the 2D-classified particles (no prior 3D cleanup).
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
September 26, 2025 at 2:28 AM
Initial attempt at replicating in relion (parameters in next post). This is for Aca2-RNA, using a 100k subset of the 2D-classified particles (no prior 3D cleanup).
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
Reposted by Jan Felix
We finally wrote up the paper for LocScale-2.0.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
We are happy to release LocScale2.0: a tool for context-aware, confidence-weighted cryoEM map optimisation.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
September 15, 2025 at 7:39 AM
We finally wrote up the paper for LocScale-2.0.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
Reposted by Jan Felix
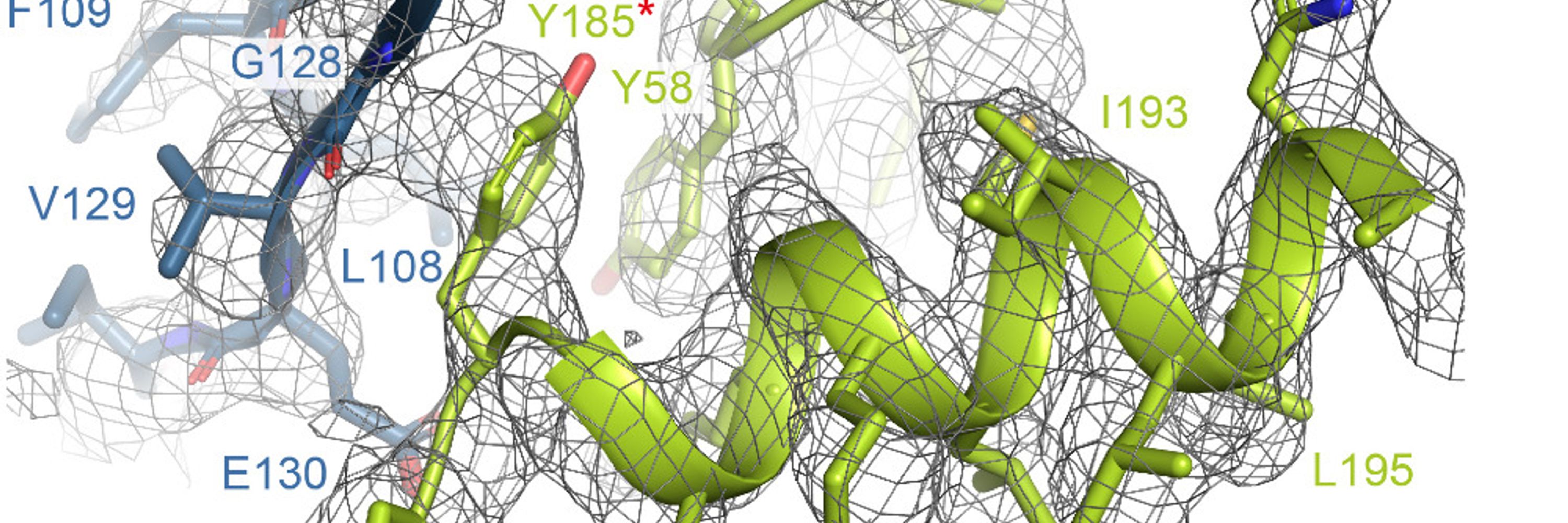
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
September 13, 2025 at 12:05 AM
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
Now published @cp-cellreports.bsky.social! Antibody-mediated TGF-β1 activation for the treatment of diseases caused by deleterious T cell activity: www.cell.com/cell-reports...
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.

September 2, 2025 at 11:25 AM
Now published @cp-cellreports.bsky.social! Antibody-mediated TGF-β1 activation for the treatment of diseases caused by deleterious T cell activity: www.cell.com/cell-reports...
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Reposted by Jan Felix
We are excited to announce the first in-cell structure of a LINC complex within a native nuclear envelope at subnanometer resolution!! #In-cell cryo-ET + #atomistic MD simulations! @tomdendooven.bsky.social et al!!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
August 5, 2025 at 12:41 PM
We are excited to announce the first in-cell structure of a LINC complex within a native nuclear envelope at subnanometer resolution!! #In-cell cryo-ET + #atomistic MD simulations! @tomdendooven.bsky.social et al!!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Finally out @natcomms.nature.com
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...

July 10, 2025 at 8:45 AM
Finally out @natcomms.nature.com
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...
Reposted by Jan Felix
If this is true, it’s abysmally shortsighted for #NIH. The new tools nurtured by the long series of #CASP challenges probably contribute far more in one year to the scientific and industrial economies than the entire funds that have supported it.
Exclusive: An international scientific competition widely credited with spurring the development of artificial intelligence for biology appears to be on its deathbed. scim.ag/44ukS90
Exclusive: Famed protein structure competition nears end as NIH grant money runs out
Agency silent on funding renewal for contest that inspired creation of AIs that predicted how proteins would fold
scim.ag
July 3, 2025 at 8:08 AM
Reposted by Jan Felix
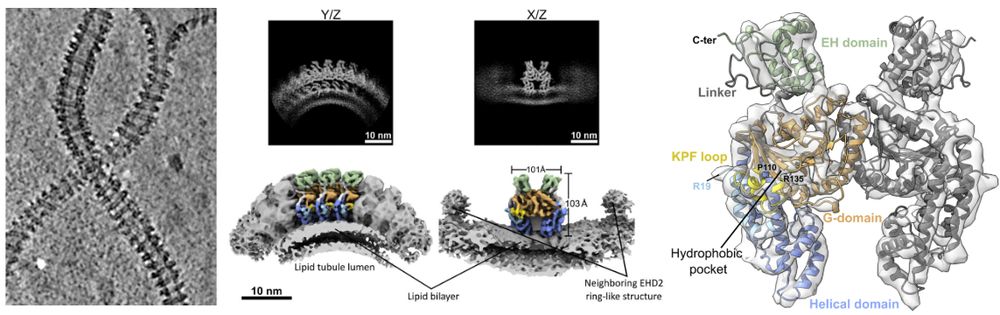
We have a preprint for you
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
June 8, 2025 at 11:22 AM
We have a preprint for you
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
Reposted by Jan Felix
I’m very happy to share my first paper on the effects of AWI interactions and flexibility on SPA data quality! Thank you to everyone on the team 🤩
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...

Untangling the effects of flexibility and the AWI in cryoEM sample preparation: A case study using KtrA
Single particle cryo-electron microscopy (cryoEM) is a powerful tool for elucidating the structures of biological macromolecules without requiring cry…
www.sciencedirect.com
June 2, 2025 at 5:18 PM
I’m very happy to share my first paper on the effects of AWI interactions and flexibility on SPA data quality! Thank you to everyone on the team 🤩
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
Reposted by Jan Felix
We are happy to release LocScale2.0: a tool for context-aware, confidence-weighted cryoEM map optimisation.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
May 22, 2025 at 10:10 AM
We are happy to release LocScale2.0: a tool for context-aware, confidence-weighted cryoEM map optimisation.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Reposted by Jan Felix
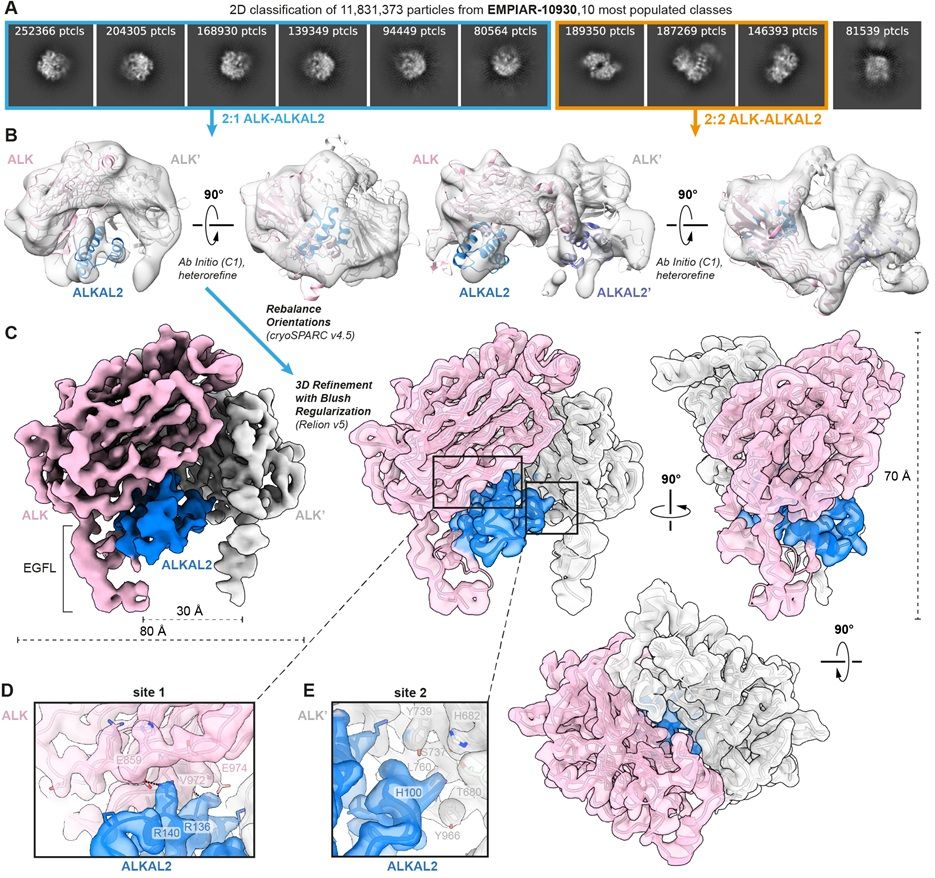
Reports of diverse stoichiometries have complicated our understanding of the complex between ALK kinase and its #cytokine ligand ALKAL2. @janfelix.bsky.social @savvideslab.bsky.social &co reanalyze #cryoEM data to clarify the situation @plosbiology.org 🧪 plos.io/4lvRfvu
April 14, 2025 at 5:34 PM
Reports of diverse stoichiometries have complicated our understanding of the complex between ALK kinase and its #cytokine ligand ALKAL2. @janfelix.bsky.social @savvideslab.bsky.social &co reanalyze #cryoEM data to clarify the situation @plosbiology.org 🧪 plos.io/4lvRfvu
Reposted by Jan Felix
Delighted to share our reanalysis of cryo-EM data in EMPIAR-10930, now at:
journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!
journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!

Reanalysis of cryo-EM data reveals ALK-cytokine assemblies with both 2:1 and 2:2 stoichiometries
The mechanistic understanding of the complex between ALK kinase and its cytokine ligand ALKAL2 has been complicated by reports of diverse stoichiometries (2:1 vs 2:2). This study reanalyzes cryo-EM da...
journals.plos.org
April 13, 2025 at 7:04 PM
Delighted to share our reanalysis of cryo-EM data in EMPIAR-10930, now at:
journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!
journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!
Reposted by Jan Felix
#CryoSPARC v4.7 is out! 🚀
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
April 9, 2025 at 2:14 PM
#CryoSPARC v4.7 is out! 🚀
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Reposted by Jan Felix
I'm super happy that our story is now published!
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
March 21, 2025 at 2:09 PM
I'm super happy that our story is now published!
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
Reposted by Jan Felix
Foam film vitrification for cryo-EM https://www.biorxiv.org/content/10.1101/2025.03.10.642366v1
March 12, 2025 at 3:50 PM
Foam film vitrification for cryo-EM https://www.biorxiv.org/content/10.1101/2025.03.10.642366v1
Reposted by Jan Felix
Structural biology is in an era of dynamics & assemblies but turning raw experimental data into atomic models at scale remains challenging. @minhuanli.bsky.social and I present ROCKET🚀: an AlphaFold augmentation that integrates crystallographic and cryoEM/ET data with room for more! 1/14.
February 24, 2025 at 12:23 PM
Structural biology is in an era of dynamics & assemblies but turning raw experimental data into atomic models at scale remains challenging. @minhuanli.bsky.social and I present ROCKET🚀: an AlphaFold augmentation that integrates crystallographic and cryoEM/ET data with room for more! 1/14.
Reposted by Jan Felix
New preprint from the @remaut-lab.bsky.social ! Happy to share our latest work on a new member of the ENA family, protein fibers found on the spore’s surface. Using CryoID @mikesleutel.bsky.social identified F-ENA in BTK. The paper dives deep into its structure analysis by integrating X-ray and AF3
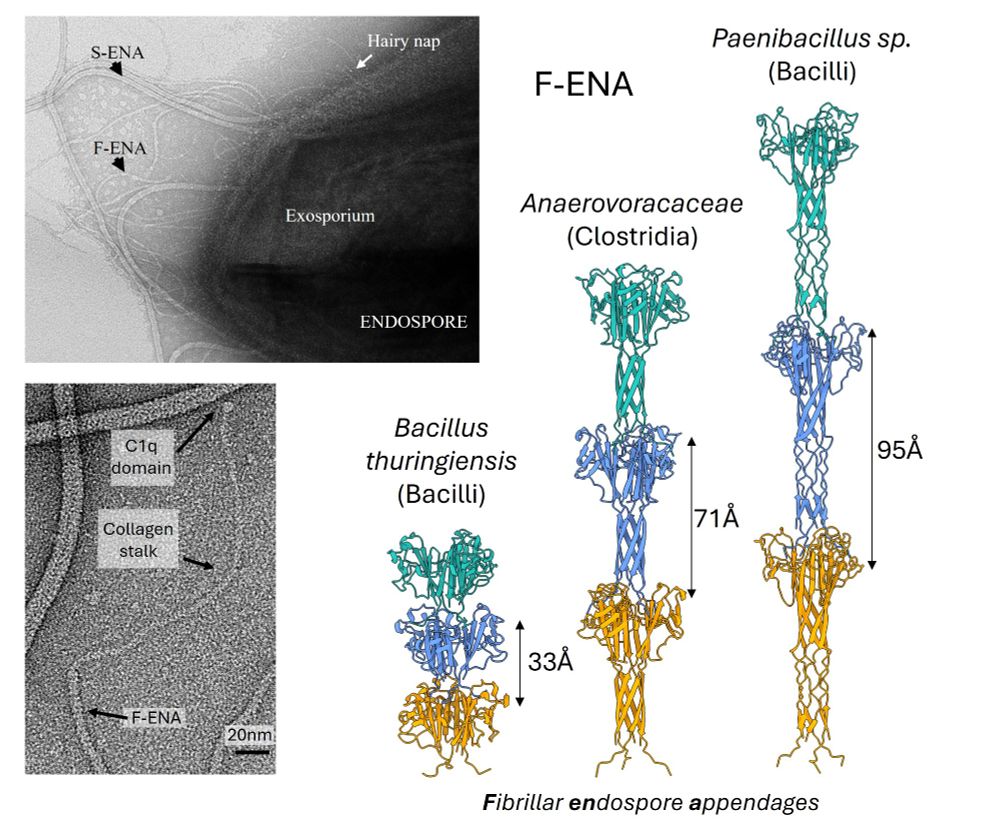
February 12, 2025 at 12:54 PM
New preprint from the @remaut-lab.bsky.social ! Happy to share our latest work on a new member of the ENA family, protein fibers found on the spore’s surface. Using CryoID @mikesleutel.bsky.social identified F-ENA in BTK. The paper dives deep into its structure analysis by integrating X-ray and AF3
Reposted by Jan Felix
Cryo-EM structures of mammalian membrane proteins from a single cell culture dish?
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.
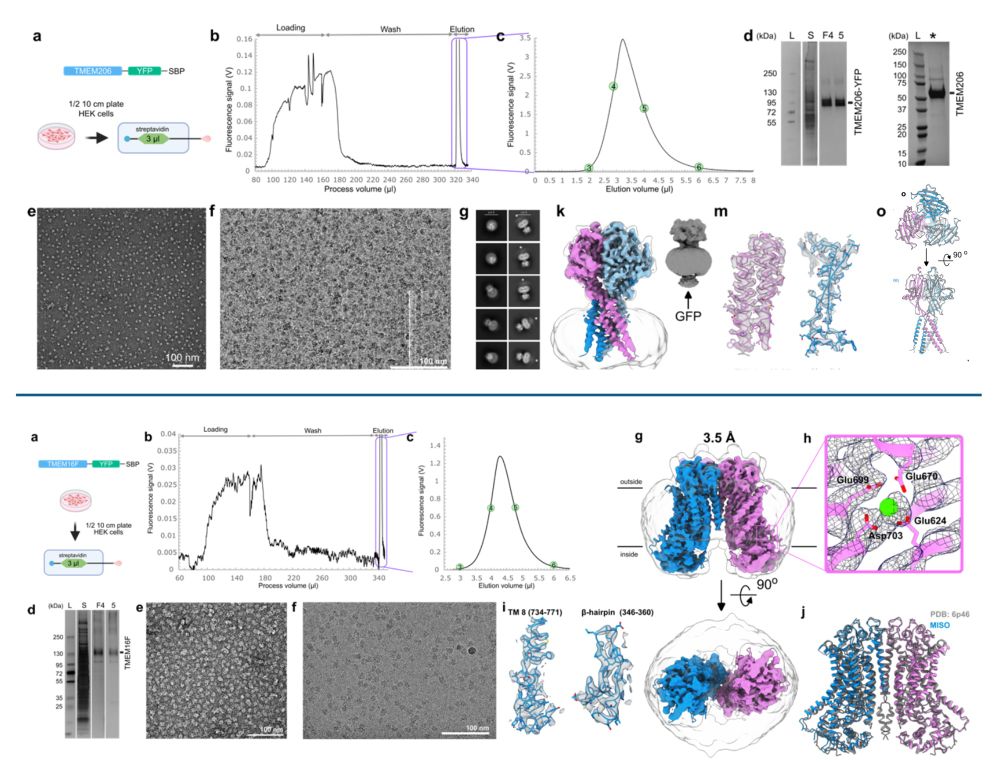
January 16, 2025 at 9:27 AM
Cryo-EM structures of mammalian membrane proteins from a single cell culture dish?
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.

