
ebi.ac.uk/interpro/



✨Updated to Pfam 38.0
✨1,068 new InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk
✨Updated to Pfam 38.0
✨1,068 new InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk



✨Updated to Pfam 38.0
✨1,068 new InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk
More info: proteinswebteam.github.io/interpro-blo...

More info: proteinswebteam.github.io/interpro-blo...
Read more here ➡️ zurl.co/poyXM
Read more here ➡️ zurl.co/poyXM
Discover the taxonomic breadth of your InterPro entry at a glance with our dynamic sunburst chart! 🕸️ 💻🧪🧬
Discover the taxonomic breadth of your InterPro entry at a glance with our dynamic sunburst chart! 🕸️ 💻🧪🧬
A Nextflow-powered, containerised overhaul for scalable, reproducible protein annotation—decoupled software/data, automated data handling, and smoother runs across HPC and cloud.
Feedback welcome!
github.com/ebi-pf-team/...
💻🧪🧬

A Nextflow-powered, containerised overhaul for scalable, reproducible protein annotation—decoupled software/data, automated data handling, and smoother runs across HPC and cloud.
Feedback welcome!
github.com/ebi-pf-team/...
💻🧪🧬
🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪



🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪
Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪

Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪
🎤 Talk 08/07 16:50, amphi E: Pfam: embracing AI/ML by @typhainepl.bsky.social
📊 Poster #97: InterPro: Accelerating Protein Annotation with AI by @matthiasblum.bsky.social
💻🧪

🎤 Talk 08/07 16:50, amphi E: Pfam: embracing AI/ML by @typhainepl.bsky.social
📊 Poster #97: InterPro: Accelerating Protein Annotation with AI by @matthiasblum.bsky.social
💻🧪
Now featuring #AlphaFold Server and AlphaFold 3. Start your training today: www.ebi.ac.uk/training/onl...
AlphaFold, developed by @ebi.embl.org and Google DeepMind.
Now featuring #AlphaFold Server and AlphaFold 3. Start your training today: www.ebi.ac.uk/training/onl...
AlphaFold, developed by @ebi.embl.org and Google DeepMind.
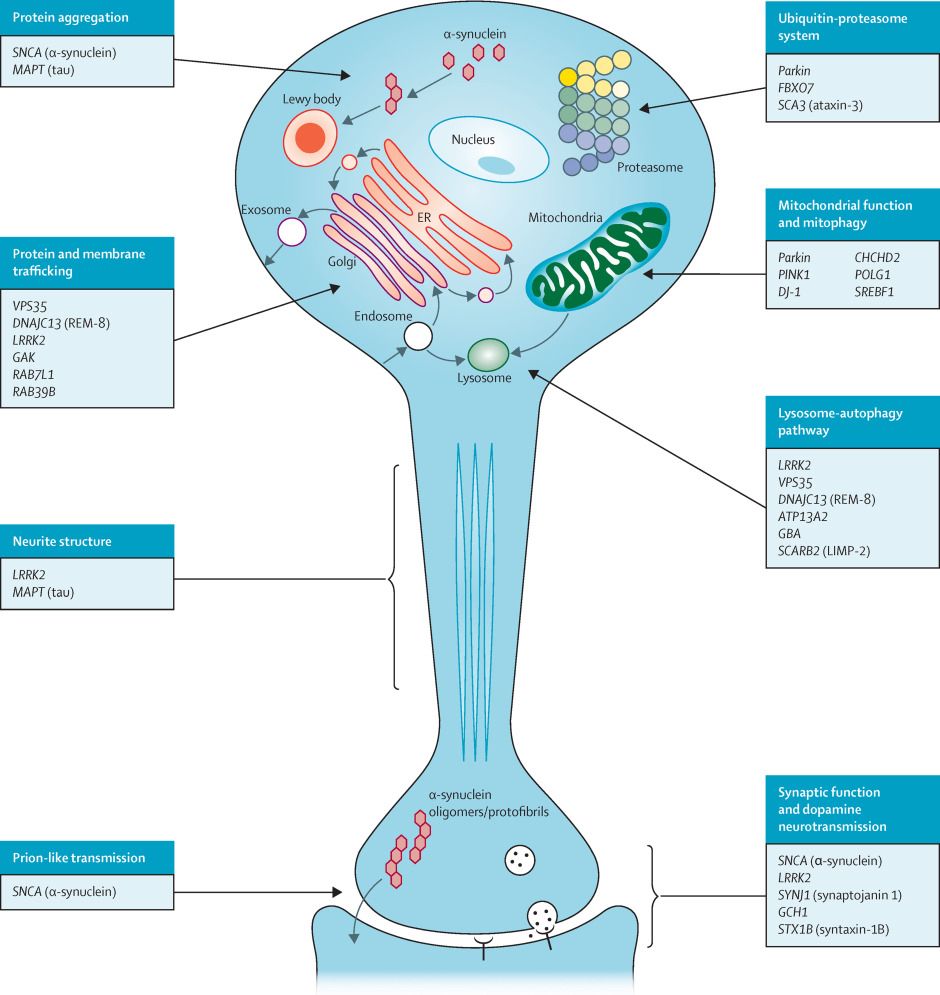
![Schematic diagram showing taste signal transmission between the tongue (with taste buds that contain taste cells harbouring GPCRs) and brain. Taste GPCRs are activated by specific food compounds, recruit specific G proteins that further induce intracellular calcium release. Adapted from [https://bio.libretexts.org/Bookshelves/Biochemistry/Fundamentals_of_Biochemistry_(Jakubowski_and_Flatt)/Unit_IV_-_Special_Topics/28:_Biosignaling_-_Capstone_Volume_I/28.18:_Signal_Transduction_-_Taste_(Gustation)].](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2b2g3csqntyhbjba6zutidvo/bafkreiarei32po5buevs63s7abazj42rdvx5w7pnszlg2zduhyqlpfqxdu@jpeg)
✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI


✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
Now with 1.8 billion AI-driven protein annotations, AlphaFold-based TED domains, and viral proteins from the Big Fantastic Virus Database (BFVD).
See it all at www.ebi.ac.uk/interpro
🖥️🧬
@interprodb.bsky.social
Now with 1.8 billion AI-driven protein annotations, AlphaFold-based TED domains, and viral proteins from the Big Fantastic Virus Database (BFVD).
See it all at www.ebi.ac.uk/interpro
🖥️🧬
@interprodb.bsky.social
For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
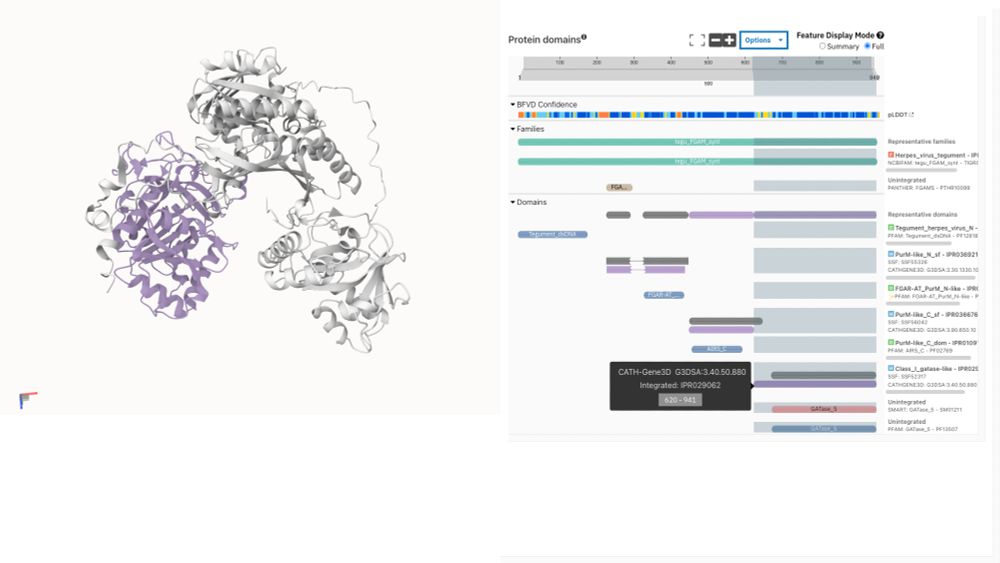
For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
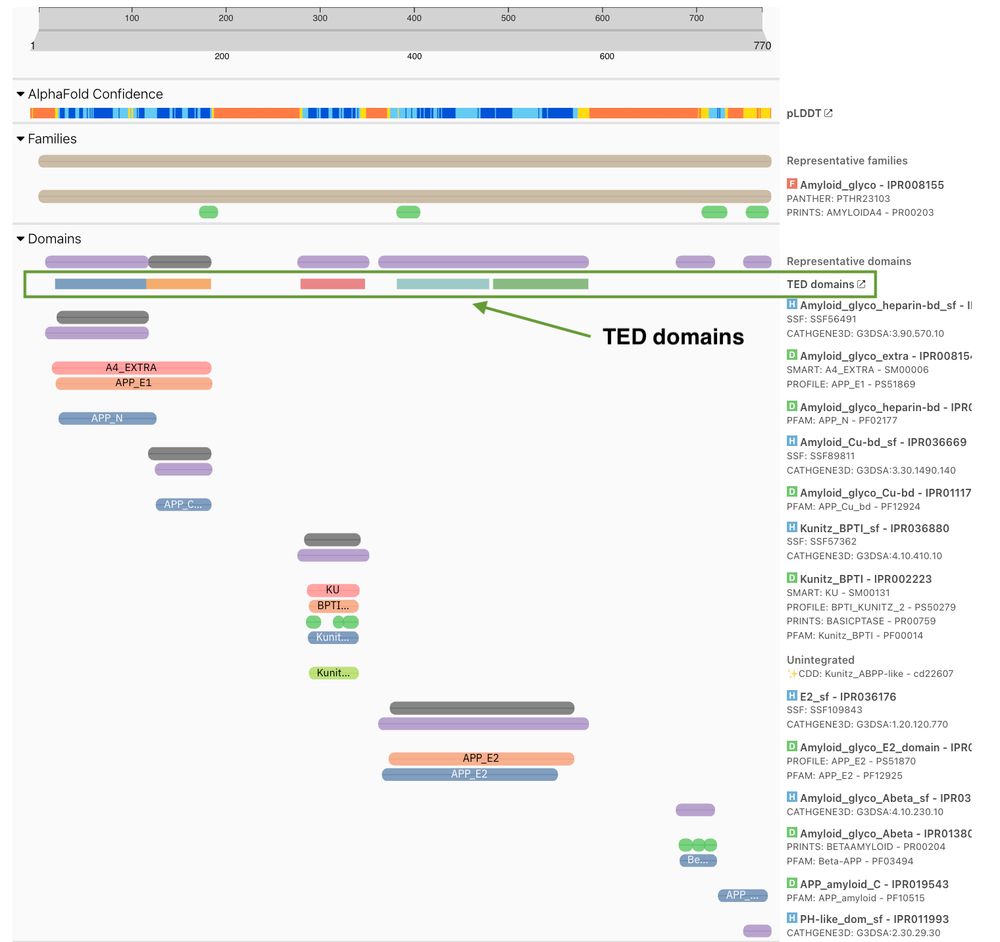
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...
✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕




✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕
@matthiasblum.bsky.social?
The recording is now available: lnkd.in/ei4fKGXd

@matthiasblum.bsky.social?
The recording is now available: lnkd.in/ei4fKGXd
🧶🧬🖥️🧪

#genomics #bioinformatics
#genomics #bioinformatics
3 ways to use it:
1️⃣ Sequence search on the InterPro website: ebi.ac.uk/interpro/sea...
2️⃣ Local installation: interproscan-docs.readthedocs.io/en/v5/Instal...
3️⃣ Local installation using a container: interproscan-docs.readthedocs.io/en/v5/HowToU...
💻🧪


3 ways to use it:
1️⃣ Sequence search on the InterPro website: ebi.ac.uk/interpro/sea...
2️⃣ Local installation: interproscan-docs.readthedocs.io/en/v5/Instal...
3️⃣ Local installation using a container: interproscan-docs.readthedocs.io/en/v5/HowToU...
💻🧪
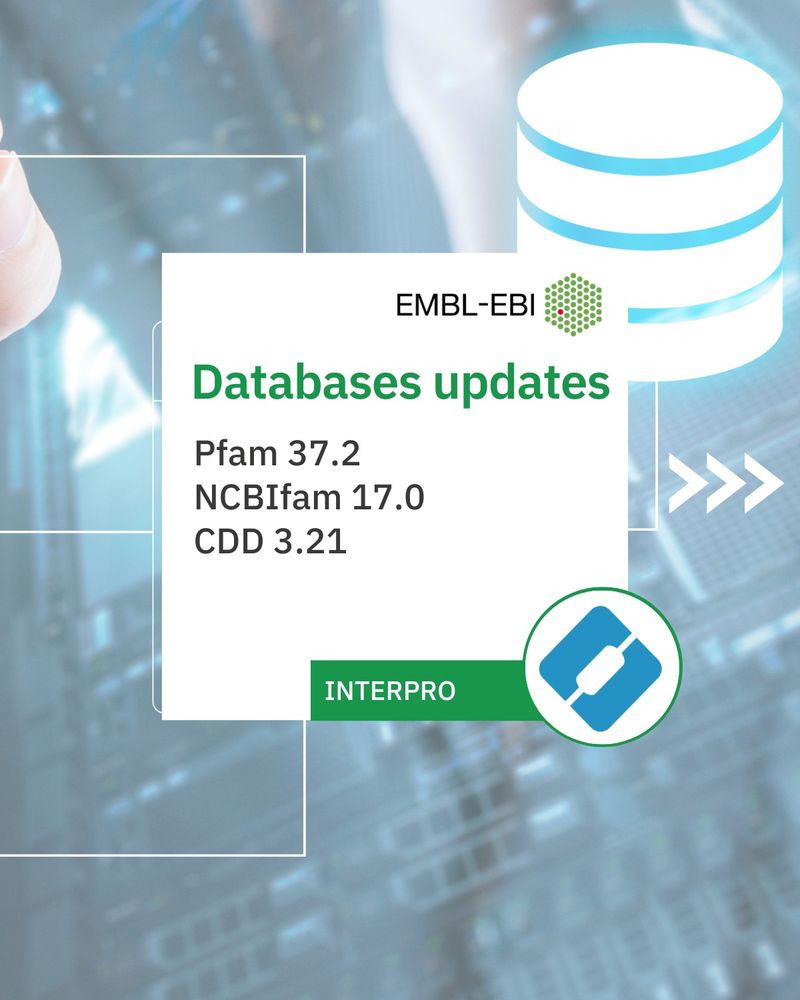
✨Update to Pfam 37.2, NCBIfam 17.0, CDD 3.21.
✨661 new entries, including 682 signatures from the Pfam (663), NCBIfam (5), PANTHER (5), CDD (9) databases. Have a look: ebi.ac.uk/interpro/ 💻🧪




✨Update to Pfam 37.2, NCBIfam 17.0, CDD 3.21.
✨661 new entries, including 682 signatures from the Pfam (663), NCBIfam (5), PANTHER (5), CDD (9) databases. Have a look: ebi.ac.uk/interpro/ 💻🧪

