
Danny IncaRNAto
@incarnatolab.bsky.social
Associate Professor in Molecular Genetics | University of Groningen | Chief R&D Officer @ Serna Bio | #RNA structural ensembles & dynamics | https://www.incaRNAtolab.com | Views are my own
Until a fix will be released, we advice users to install ViennaRNA v2.7.0 in a separate folder and to provide the path to RNAplot v2.7.0 via the -vrp parameter of rf-fold. (4/n)
October 13, 2025 at 12:47 PM
Until a fix will be released, we advice users to install ViennaRNA v2.7.0 in a separate folder and to provide the path to RNAplot v2.7.0 via the -vrp parameter of rf-fold. (4/n)
!!! Important note: A bug was introduced in ViennaRNA v2.7.0, which breaks the pseudoknot detection functionality of rf-fold. However, RNAplot v2.7.0 is required to take advantage of the new secondary structure plotting functionality. (3/n)
October 13, 2025 at 12:47 PM
!!! Important note: A bug was introduced in ViennaRNA v2.7.0, which breaks the pseudoknot detection functionality of rf-fold. However, RNAplot v2.7.0 is required to take advantage of the new secondary structure plotting functionality. (3/n)
Highlights: new normalization method in rf-norm (Mitchell method, pubmed.ncbi.nlm.nih.gov/37334863/), statistics plots in rf-count & rf-count-genome, and secondary structure plots in rf-fold (powered by ViennaRNA v2.7.0) (2/n)

October 13, 2025 at 12:47 PM
Highlights: new normalization method in rf-norm (Mitchell method, pubmed.ncbi.nlm.nih.gov/37334863/), statistics plots in rf-count & rf-count-genome, and secondary structure plots in rf-fold (powered by ViennaRNA v2.7.0) (2/n)
Thanks Lars! It definitely was :-)
July 30, 2025 at 8:47 AM
Thanks Lars! It definitely was :-)
Thanks Dirk-Jan!
July 29, 2025 at 4:54 PM
Thanks Dirk-Jan!
Thanks Sebastian!
July 27, 2025 at 8:11 PM
Thanks Sebastian!
And a huge thanks to @erc.europa.eu for funding that made all of this possible! (11/n)
July 27, 2025 at 5:03 PM
And a huge thanks to @erc.europa.eu for funding that made all of this possible! (11/n)
Shoutout to rockstar Ivana Borovská, who led this incredible work, and to our collaborators, particularly @mtwolfinger.bsky.social and Wim Velema! (10/n)
July 27, 2025 at 5:03 PM
Shoutout to rockstar Ivana Borovská, who led this incredible work, and to our collaborators, particularly @mtwolfinger.bsky.social and Wim Velema! (10/n)
While our study still presents many limitations, we sincerely hope this will represent a stepping stone in better understanding and characterizing the structural dynamics of cellular RNAs, and their roles in regulating gene expression. (9/n)
July 27, 2025 at 5:03 PM
While our study still presents many limitations, we sincerely hope this will represent a stepping stone in better understanding and characterizing the structural dynamics of cellular RNAs, and their roles in regulating gene expression. (9/n)
Furthermore, we introduced the DeConStruct framework (github.com/dincarnato/p...), to aid the discovery and prioritization of conserved regulatory RNA structural switches discovered via ensemble deconvolution analysis of chemical probing data. (8/n)
papers/Borovska et al., 2025 at main · dincarnato/papers
Data & pipelines associated with papers from the lab - dincarnato/papers
github.com
July 27, 2025 at 5:03 PM
Furthermore, we introduced the DeConStruct framework (github.com/dincarnato/p...), to aid the discovery and prioritization of conserved regulatory RNA structural switches discovered via ensemble deconvolution analysis of chemical probing data. (8/n)
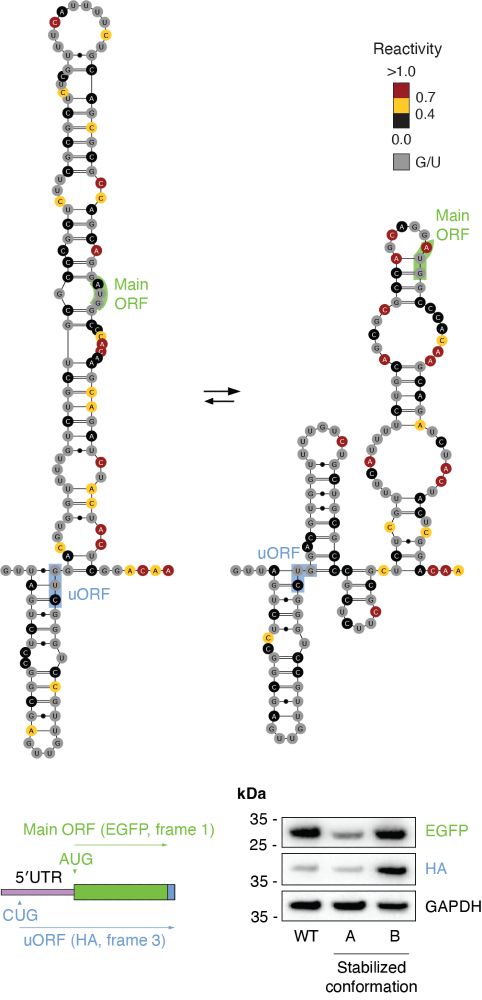
For example, the 5′-UTR of the CKS2 RNA populates two conformations, one promoting increased translation of a uORF, and one reducing translation of the main ORF. (7/n)
July 27, 2025 at 5:03 PM
For example, the 5′-UTR of the CKS2 RNA populates two conformations, one promoting increased translation of a uORF, and one reducing translation of the main ORF. (7/n)
Thanks to this method, we observed that regions of human 5′-UTRs populating 2+ alternative conformations, tend to be enriched for upstream start codons (NTG), suggesting that the switch between these conformations might regulate translation of uORFs vs. main ORF. (6/n)
July 27, 2025 at 5:03 PM
Thanks to this method, we observed that regions of human 5′-UTRs populating 2+ alternative conformations, tend to be enriched for upstream start codons (NTG), suggesting that the switch between these conformations might regulate translation of uORFs vs. main ORF. (6/n)
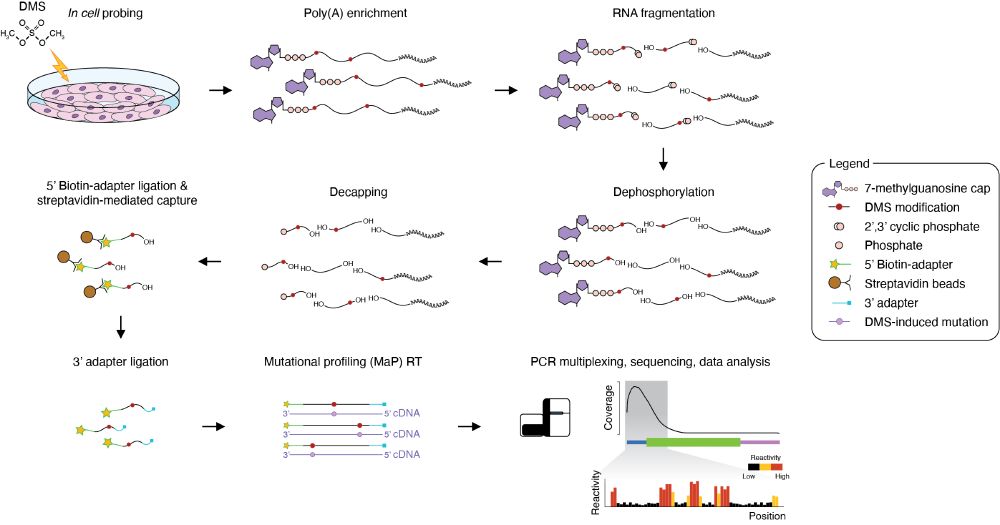
Next, we sought to explore the transcriptome of human cells. Due to the extreme sequencing depth required for ensemble deconvolution analyses, we developed 5′UTR-MaP, which enables the transcriptome-scale interrogation of 5′-UTRs (and, in general, of 5′-terminal regions). (5/n)
July 27, 2025 at 5:03 PM
Next, we sought to explore the transcriptome of human cells. Due to the extreme sequencing depth required for ensemble deconvolution analyses, we developed 5′UTR-MaP, which enables the transcriptome-scale interrogation of 5′-UTRs (and, in general, of 5′-terminal regions). (5/n)
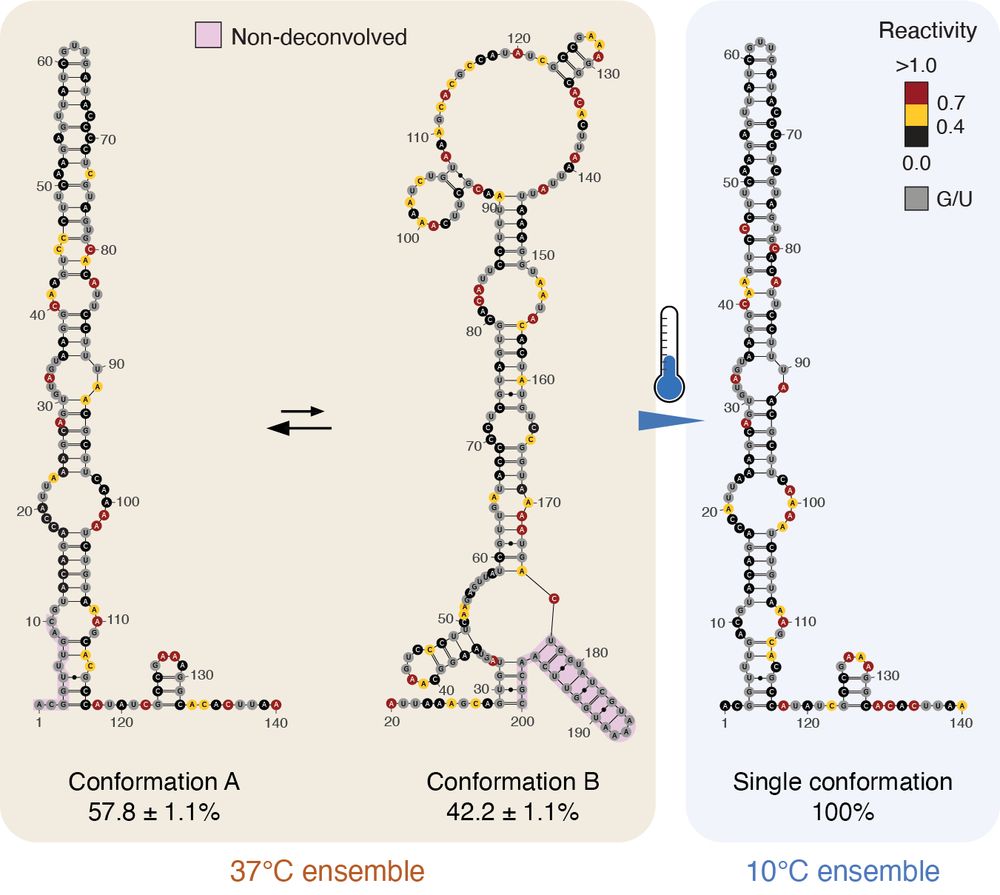
Interestingly, the well characterized cspA thermometer already populates both ON and OFF conformations at 37°C, and completely shifts to the ON conformation at 10°C, explaining why cspA is among the top-expressed proteins in the E. coli proteome. (4/n)
July 27, 2025 at 5:03 PM
Interestingly, the well characterized cspA thermometer already populates both ON and OFF conformations at 37°C, and completely shifts to the ON conformation at 10°C, explaining why cspA is among the top-expressed proteins in the E. coli proteome. (4/n)
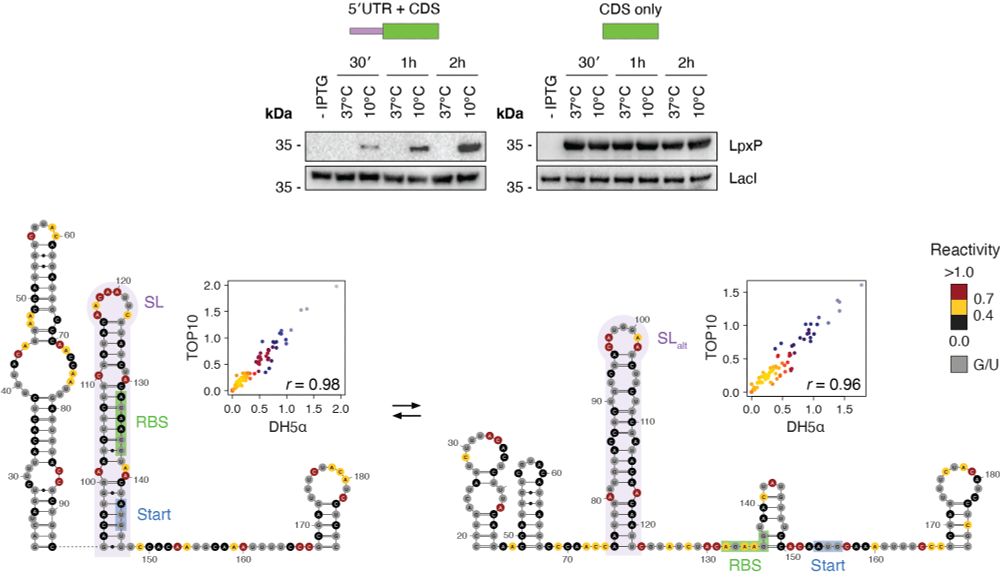
Subjecting bacteria to cold shock causes an increase in the number of regions populating 2+ alternative conformations. By combining these results with covariation analysis we identified a number of novel RNA cold thermometers, such as the 5′ UTR of lpxP. (3/n)
July 27, 2025 at 5:03 PM
Subjecting bacteria to cold shock causes an increase in the number of regions populating 2+ alternative conformations. By combining these results with covariation analysis we identified a number of novel RNA cold thermometers, such as the 5′ UTR of lpxP. (3/n)

