rdcu.be/exqtH
Lastly, this wouldn't have been possible without a fantastic team of co-authors ☺️ Truly grateful to everyone who contributed and supported the work along the way.
#BaseEditing #PrecisionOncology #TP53 #CRISPR
rdcu.be/exqtH
Lastly, this wouldn't have been possible without a fantastic team of co-authors ☺️ Truly grateful to everyone who contributed and supported the work along the way.
#BaseEditing #PrecisionOncology #TP53 #CRISPR
✅ Base editing can reverse oncogenic phenotypes
✅ Repairing TP53 hotspots restores a conserved p53 regulatory network
✅ Our platform is applicable to other mutations
✅ Precision editing + transcriptomics = powerful combo
✅ Base editing can reverse oncogenic phenotypes
✅ Repairing TP53 hotspots restores a conserved p53 regulatory network
✅ Our platform is applicable to other mutations
✅ Precision editing + transcriptomics = powerful combo
🏥 Patient-derived organoids
🧬 Multi-omics profiling
🧫 Single cells profiling
Our platform opens the door to a deeper, mutation-level understanding of cancer dependencies and vulnerabilities.
🏥 Patient-derived organoids
🧬 Multi-omics profiling
🧫 Single cells profiling
Our platform opens the door to a deeper, mutation-level understanding of cancer dependencies and vulnerabilities.
It’s a step toward precision oncology grounded in functional genetics - not just correlational genomics.
We now have a way to test which mutations matter and how they reshape cellular programs.
It’s a step toward precision oncology grounded in functional genetics - not just correlational genomics.
We now have a way to test which mutations matter and how they reshape cellular programs.
🧬 Correct mutations
📊 Measure downstream gene expression
💡 Reveal functional consequences
All in endogenous contexts.

🧬 Correct mutations
📊 Measure downstream gene expression
💡 Reveal functional consequences
All in endogenous contexts.
Repairing mutations in KRAS, PTEN or SMAD4, we found gene-specific differences in how mutations drive cancer cell fitness
Reinstating brakes (TP53) seems more effective than releasing a pedal stuck on gas (KRAS)


Repairing mutations in KRAS, PTEN or SMAD4, we found gene-specific differences in how mutations drive cancer cell fitness
Reinstating brakes (TP53) seems more effective than releasing a pedal stuck on gas (KRAS)
✔️ Consistent across tissue types
✔️ Independent of co-occurring mutations
✔️ Uniform across TP53 mutations (R273H vs R175H)
That suggests a conserved, mutation-independent p53 regulatory network that controls cancer cells. 🧬🛡️

✔️ Consistent across tissue types
✔️ Independent of co-occurring mutations
✔️ Uniform across TP53 mutations (R273H vs R175H)
That suggests a conserved, mutation-independent p53 regulatory network that controls cancer cells. 🧬🛡️
🎯 Cancer cells are dependent on the mutant TP53 allele for survival.
When we corrected these mutations, we saw:
✅ Loss of oncogenic proliferation
✅ Reactivation of tumor suppressive transcriptional programs


🎯 Cancer cells are dependent on the mutant TP53 allele for survival.
When we corrected these mutations, we saw:
✅ Loss of oncogenic proliferation
✅ Reactivation of tumor suppressive transcriptional programs
No overexpression. No exogenous DNA. Just editing the native genome.
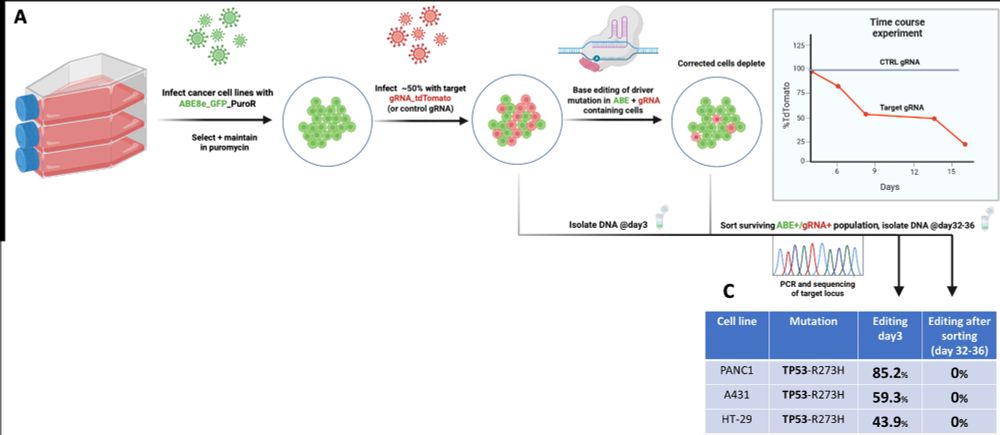
No overexpression. No exogenous DNA. Just editing the native genome.

