🧬 Correct mutations
📊 Measure downstream gene expression
💡 Reveal functional consequences
All in endogenous contexts.

🧬 Correct mutations
📊 Measure downstream gene expression
💡 Reveal functional consequences
All in endogenous contexts.
Repairing mutations in KRAS, PTEN or SMAD4, we found gene-specific differences in how mutations drive cancer cell fitness
Reinstating brakes (TP53) seems more effective than releasing a pedal stuck on gas (KRAS)


Repairing mutations in KRAS, PTEN or SMAD4, we found gene-specific differences in how mutations drive cancer cell fitness
Reinstating brakes (TP53) seems more effective than releasing a pedal stuck on gas (KRAS)
✔️ Consistent across tissue types
✔️ Independent of co-occurring mutations
✔️ Uniform across TP53 mutations (R273H vs R175H)
That suggests a conserved, mutation-independent p53 regulatory network that controls cancer cells. 🧬🛡️

✔️ Consistent across tissue types
✔️ Independent of co-occurring mutations
✔️ Uniform across TP53 mutations (R273H vs R175H)
That suggests a conserved, mutation-independent p53 regulatory network that controls cancer cells. 🧬🛡️
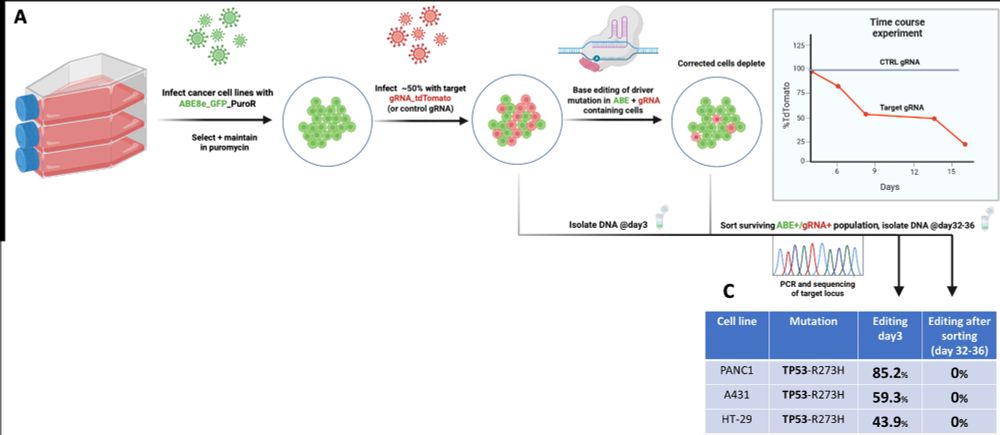
🎯 Cancer cells are dependent on the mutant TP53 allele for survival.
When we corrected these mutations, we saw:
✅ Loss of oncogenic proliferation
✅ Reactivation of tumor suppressive transcriptional programs


🎯 Cancer cells are dependent on the mutant TP53 allele for survival.
When we corrected these mutations, we saw:
✅ Loss of oncogenic proliferation
✅ Reactivation of tumor suppressive transcriptional programs
No overexpression. No exogenous DNA. Just editing the native genome.
No overexpression. No exogenous DNA. Just editing the native genome.
We present a base editing platform to functionally & transcriptionally profile cancer hotspot mutations, starting with TP53 - the most frequently mutated gene in cancer. Let's dive in 👇🧵

We present a base editing platform to functionally & transcriptionally profile cancer hotspot mutations, starting with TP53 - the most frequently mutated gene in cancer. Let's dive in 👇🧵

