
Ulrich Hohmann
@hohmannulrich.bsky.social
Thanks a lot Yasin!
October 13, 2025 at 7:54 PM
Thanks a lot Yasin!
Shout-out also to my wonderful colleagues in the Hothorn / Brennecke / Plaschka labs, as well as to the great environment and many colleagues at the University of Geneva, @viennabiocenter.bsky.social, @impvienna.bsky.social, @imbavienna.bsky.social
October 12, 2025 at 8:19 PM
Shout-out also to my wonderful colleagues in the Hothorn / Brennecke / Plaschka labs, as well as to the great environment and many colleagues at the University of Geneva, @viennabiocenter.bsky.social, @impvienna.bsky.social, @imbavienna.bsky.social
This would have been impossible without the amazing support from my PhD mentor @structplantbio.bsky.social, and my postdoc mentors @juliusbrennecke.bsky.social and Clemens Plaschka (@plaschkalab.bsky.social)!
October 12, 2025 at 8:19 PM
This would have been impossible without the amazing support from my PhD mentor @structplantbio.bsky.social, and my postdoc mentors @juliusbrennecke.bsky.social and Clemens Plaschka (@plaschkalab.bsky.social)!
Apply to the PhD program until Oct. 16th: www.imb.de/students-pos...
More info on our future lab:
www.imb.de/research/our...
I’m looking forward to future endeavours in Mainz and exciting science to happen!
More info on our future lab:
www.imb.de/research/our...
I’m looking forward to future endeavours in Mainz and exciting science to happen!
Apply to the IPP
IMB Mainz
www.imb.de
October 12, 2025 at 8:19 PM
Apply to the PhD program until Oct. 16th: www.imb.de/students-pos...
More info on our future lab:
www.imb.de/research/our...
I’m looking forward to future endeavours in Mainz and exciting science to happen!
More info on our future lab:
www.imb.de/research/our...
I’m looking forward to future endeavours in Mainz and exciting science to happen!
While you’re here: also have a look at the great related work on LENG8 from YongshengShi’s lab: www.biorxiv.org/content/10.1...

LENG8 mediates RNA nuclear retention and degradation in eukaryotes
In eukaryotes, incompletely processed and misprocessed mRNAs as well as numerous noncoding RNAs are retained in the nucleus and often degraded. However, the mechanisms for this critical quality contro...
www.biorxiv.org
September 22, 2025 at 11:23 PM
While you’re here: also have a look at the great related work on LENG8 from YongshengShi’s lab: www.biorxiv.org/content/10.1...
Wonderful collaboration with @max2max.bsky.social in Clemen’s lab, @abugai.bsky.social and @lorenzoana.bsky.social in Torben’s lab. Impossible without the support from Plaschka/ Brennecke/ Jensen labs @viennabiocenter.bsky.social, @imbavienna.bsky.social, @impvienna.bsky.social (10/10)
September 22, 2025 at 11:23 PM
Wonderful collaboration with @max2max.bsky.social in Clemen’s lab, @abugai.bsky.social and @lorenzoana.bsky.social in Torben’s lab. Impossible without the support from Plaschka/ Brennecke/ Jensen labs @viennabiocenter.bsky.social, @imbavienna.bsky.social, @impvienna.bsky.social (10/10)
In sum our work reveals that (m)RNA-clamped UAP56 not only promotes mRNA export by binding TREX-2 at the NPC. It can also be read out by the LENG8-PS module in PAXT to destine the RNA for decay. (9/x)

September 22, 2025 at 11:23 PM
In sum our work reveals that (m)RNA-clamped UAP56 not only promotes mRNA export by binding TREX-2 at the NPC. It can also be read out by the LENG8-PS module in PAXT to destine the RNA for decay. (9/x)
Perturbation of the LENG8–ZFC3H1 interface, or rapid depletion of either protein, leads to the upregulation of bona fide PAXT polyA-RNA targets. This demonstrates that the LENG8-PS module is functionally important for PAXT function. (8/x)

September 22, 2025 at 11:23 PM
Perturbation of the LENG8–ZFC3H1 interface, or rapid depletion of either protein, leads to the upregulation of bona fide PAXT polyA-RNA targets. This demonstrates that the LENG8-PS module is functionally important for PAXT function. (8/x)
Alphafold reveals a conserved mode of interaction between LENG8 and the PAXT scaffolding subunit ZFC3H1, which we can validate in vivo. Thus, LENG8-PS constitutes a TREX-2-like module of PAXT. (7/x)

September 22, 2025 at 11:23 PM
Alphafold reveals a conserved mode of interaction between LENG8 and the PAXT scaffolding subunit ZFC3H1, which we can validate in vivo. Thus, LENG8-PS constitutes a TREX-2-like module of PAXT. (7/x)
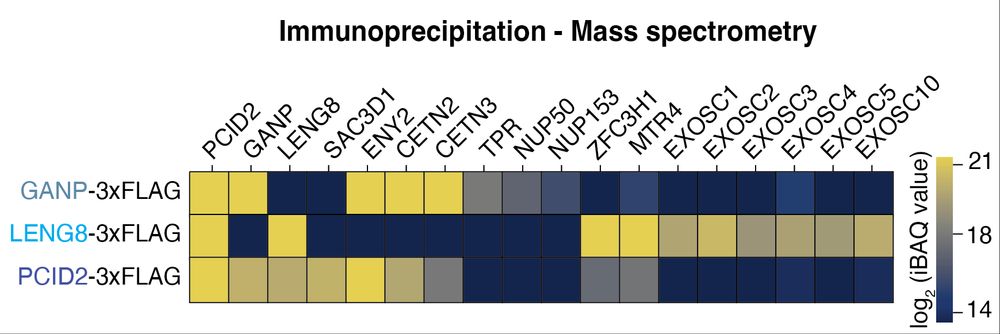
To our great surprise we found that LENG8-PS links to the ′Poly(A) tail exosome targeting (PAXT)’ connection, implicated in the degradation of polyadenylated nuclear RNA through the exosome. (6/x)
September 22, 2025 at 11:23 PM
To our great surprise we found that LENG8-PS links to the ′Poly(A) tail exosome targeting (PAXT)’ connection, implicated in the degradation of polyadenylated nuclear RNA through the exosome. (6/x)
Turns out they do! Both additional complexes bind UAP56 in vitro, and, like TREX-2, stimulate the release of RNA from UAP56. Seems counter-productive for mRNA export. So, what is their function? (5/x)

September 22, 2025 at 11:23 PM
Turns out they do! Both additional complexes bind UAP56 in vitro, and, like TREX-2, stimulate the release of RNA from UAP56. Seems counter-productive for mRNA export. So, what is their function? (5/x)
Most eukaryotes harbor two additional, less characterized TREX-2 like complexes: LENG8-PS and SAC3D1-PS, which interestingly differ in their subcellular localization. What is their function, and do they also act on UAP56? (4/x)

September 22, 2025 at 11:23 PM
Most eukaryotes harbor two additional, less characterized TREX-2 like complexes: LENG8-PS and SAC3D1-PS, which interestingly differ in their subcellular localization. What is their function, and do they also act on UAP56? (4/x)
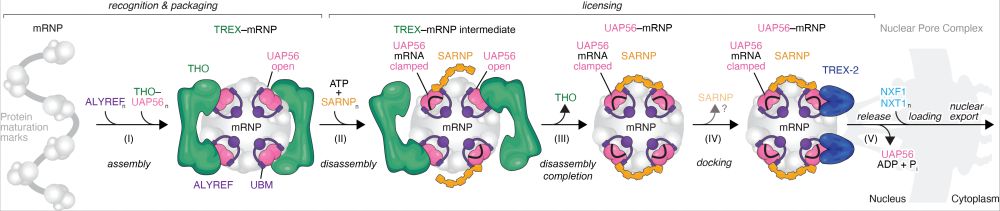
In the final step of mRNA export UAP56 enables docking of the mRNP (mRNA+bound proteins) to TREX-2, which is anchored at the nuclear pore. TREX-2 in turn removes UAP56 from the mRNA, enabling the release of the mRNP from the nuclear pore and its shuttling to the cytoplasm. (3/x)

September 22, 2025 at 11:23 PM
In the final step of mRNA export UAP56 enables docking of the mRNP (mRNA+bound proteins) to TREX-2, which is anchored at the nuclear pore. TREX-2 in turn removes UAP56 from the mRNA, enabling the release of the mRNP from the nuclear pore and its shuttling to the cytoplasm. (3/x)
We previously suggested a model for a core mRNA nuclear export pathway, which centers on the action of the RNA clampase UAP56: www.biorxiv.org/content/10.1... (2/x)
September 22, 2025 at 11:23 PM
We previously suggested a model for a core mRNA nuclear export pathway, which centers on the action of the RNA clampase UAP56: www.biorxiv.org/content/10.1... (2/x)

