building models and software in genome biology.
3D genome structure in mitosis | DNA repair | meiosis.
A group leader at @IMBA_Vienna.
Dad x2.



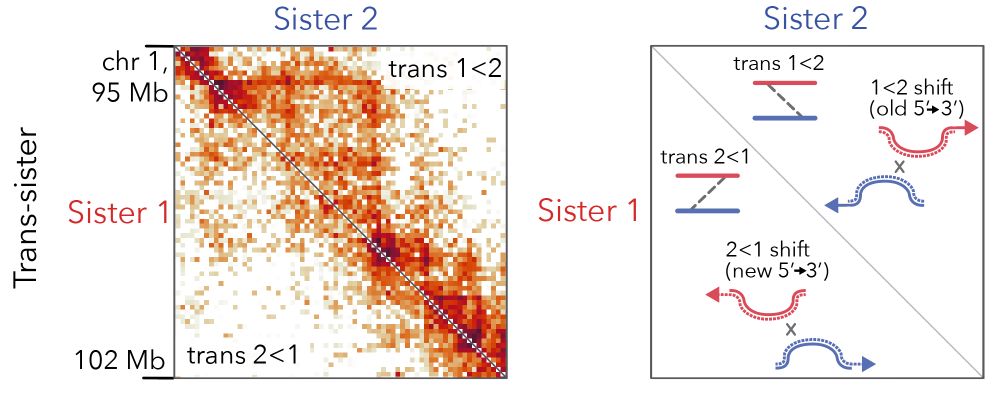
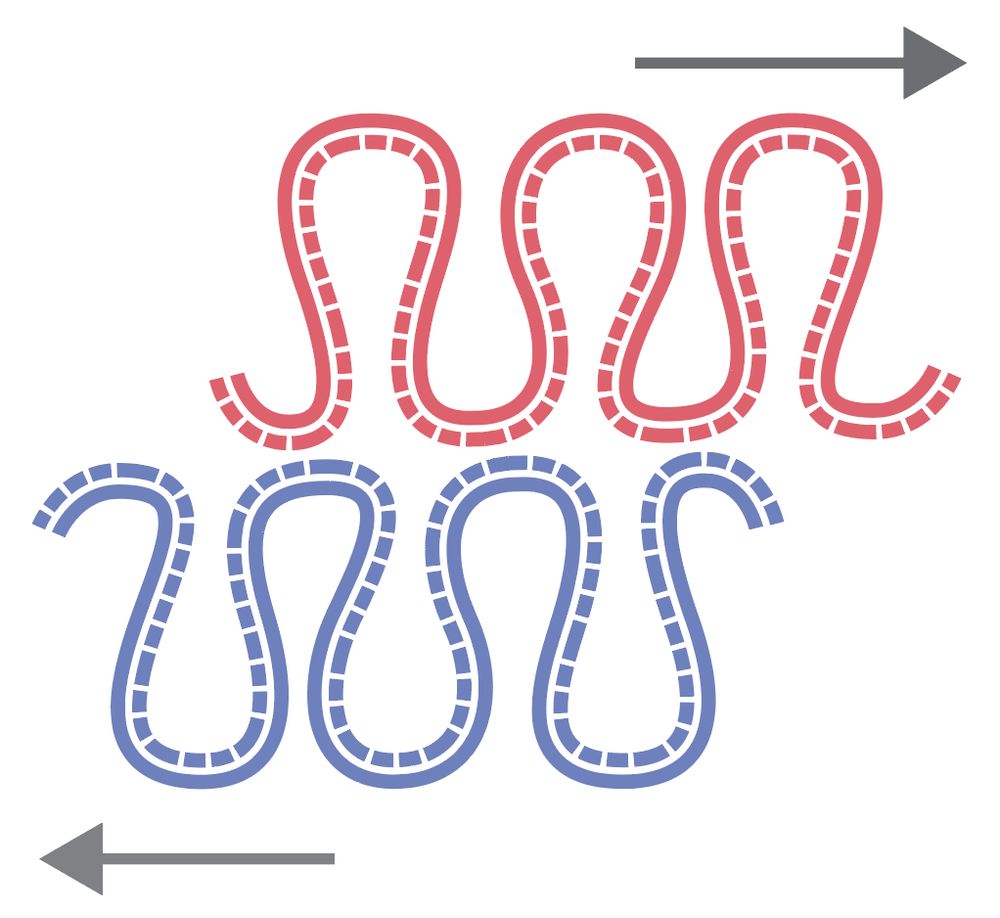
Model 1: cohesins are pushed/dragged by forks along the leading strand while staying put on the lagging strand, generating the offset. 11/

Model 1: cohesins are pushed/dragged by forks along the leading strand while staying put on the lagging strand, generating the offset. 11/








www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/


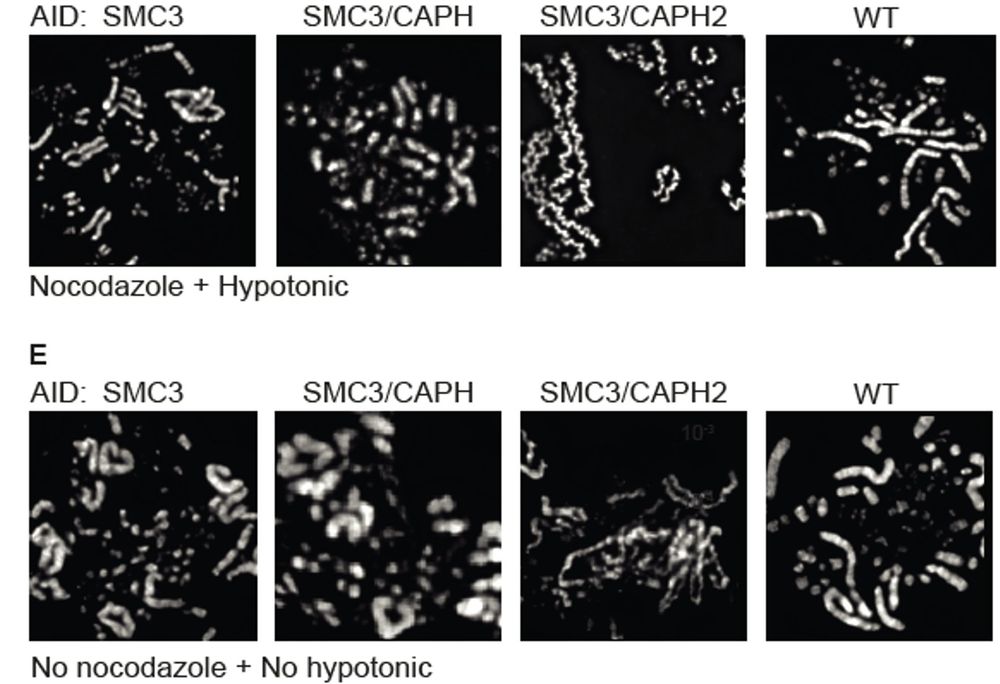
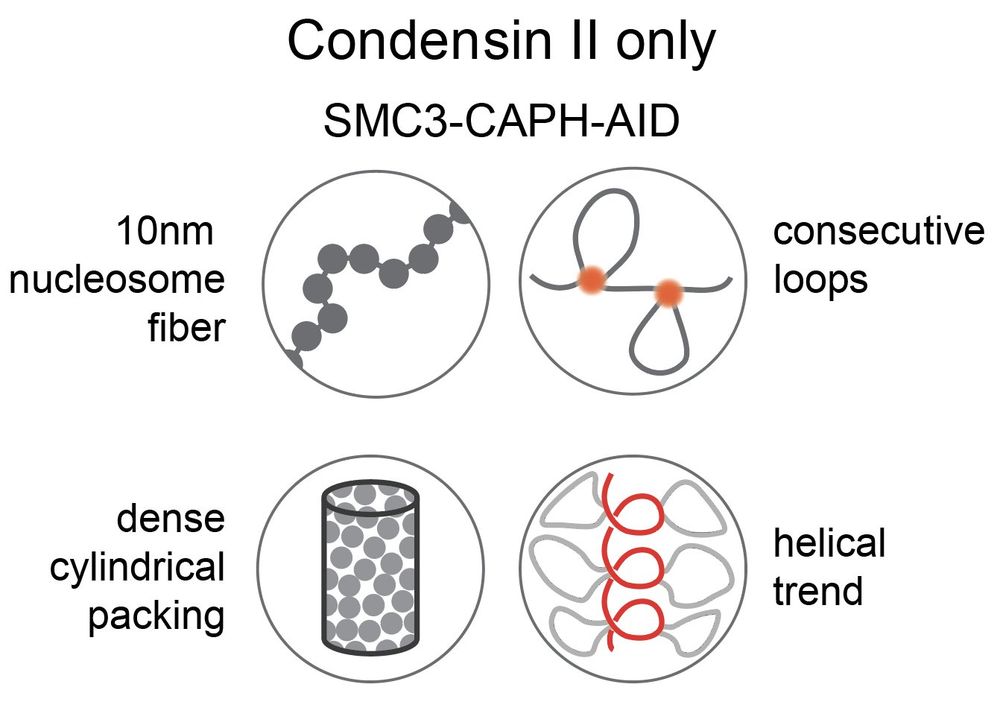
(i) condensin II scaffold is discontinuous with gaps between loops
(ii) strong second diagonals in population Hi-C can be explained by weak, irregular condensin II-mediated spiraling consistent with microscopy data

(i) condensin II scaffold is discontinuous with gaps between loops
(ii) strong second diagonals in population Hi-C can be explained by weak, irregular condensin II-mediated spiraling consistent with microscopy data