
Yohsuke T. Fukai
@fukaity.bsky.social
Postdoctoral Researcher at BDR, RIKEN
Nonequilibrium physics of living matter Riken Hakubi research team.
Twitter: https://twitter.com/fukaity
HP: https://yfukai.net/
Nonequilibrium physics of living matter Riken Hakubi research team.
Twitter: https://twitter.com/fukaity
HP: https://yfukai.net/
Community information is here napari.org/stable/commu...

Community
There are several different ways to be a part of the napari community. From being a code or documentation contributor, to creating educational content or examples, you are welcome to help develop and ...
napari.org
October 31, 2025 at 3:17 AM
Community information is here napari.org/stable/commu...
We’re entering an exciting era of gene-scale chromatin reconstitution with precise modification patterns🚀 We hope this work will stimulate further studies to understand how different histone modifications and solution environment control chromatin architecture and function. 5/5
January 5, 2025 at 9:12 AM
We’re entering an exciting era of gene-scale chromatin reconstitution with precise modification patterns🚀 We hope this work will stimulate further studies to understand how different histone modifications and solution environment control chromatin architecture and function. 5/5
We developed an in vitro Hi-C method to map nucleosome interactions and found how the acetylation reduces the nucleosome interaction frequency. For the ligated arrays (12-96-mer mixture), we found that the acetylation patterns directly control the long-range contact patterns. 4/5




January 5, 2025 at 9:12 AM
We developed an in vitro Hi-C method to map nucleosome interactions and found how the acetylation reduces the nucleosome interaction frequency. For the ligated arrays (12-96-mer mixture), we found that the acetylation patterns directly control the long-range contact patterns. 4/5
We observed arrays with different H4 hyperacetylation patterns, and found that higher acetylation density makes the array larger. There's an interesting relationship between the size and fluctuation timescales which agrees with the prediction of a simple polymer model. 3/5




January 5, 2025 at 9:12 AM
We observed arrays with different H4 hyperacetylation patterns, and found that higher acetylation density makes the array larger. There's an interesting relationship between the size and fluctuation timescales which agrees with the prediction of a simple polymer model. 3/5
How do histone modifications affect large-scale chromatin structure? We tackled this question in vitro by reconstituting gene-scale chromatin arrays with precise histone modification patterns and analyzing them using single-molecule fluorescence imaging and in vitro Hi-C. 2/5

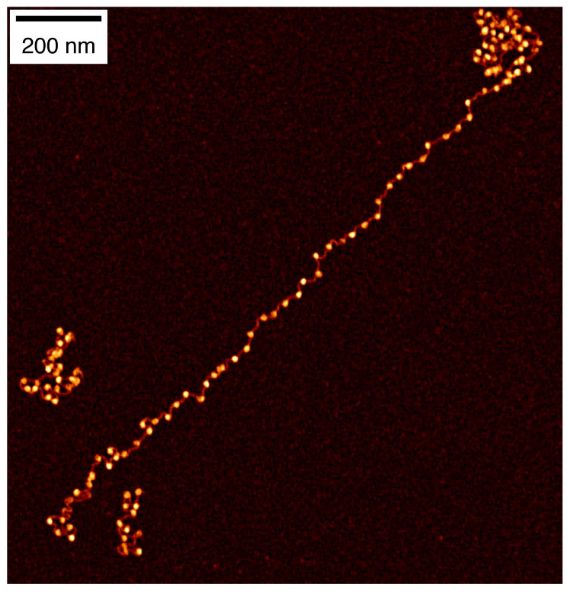
January 5, 2025 at 9:12 AM
How do histone modifications affect large-scale chromatin structure? We tackled this question in vitro by reconstituting gene-scale chromatin arrays with precise histone modification patterns and analyzing them using single-molecule fluorescence imaging and in vitro Hi-C. 2/5

