
You can access the article with an academic/non-profit registration.
You can access the article with an academic/non-profit registration.
Die Studie wurde in @natcomms.nature.com veröffentlicht👇
www.nature.com/articles/s41...
Die Studie wurde in @natcomms.nature.com veröffentlicht👇
www.nature.com/articles/s41...
www.uni-koeln.de/universitaet...
www.uni-koeln.de/en/universit... (6/7)
www.uni-koeln.de/universitaet...
www.uni-koeln.de/en/universit... (6/7)
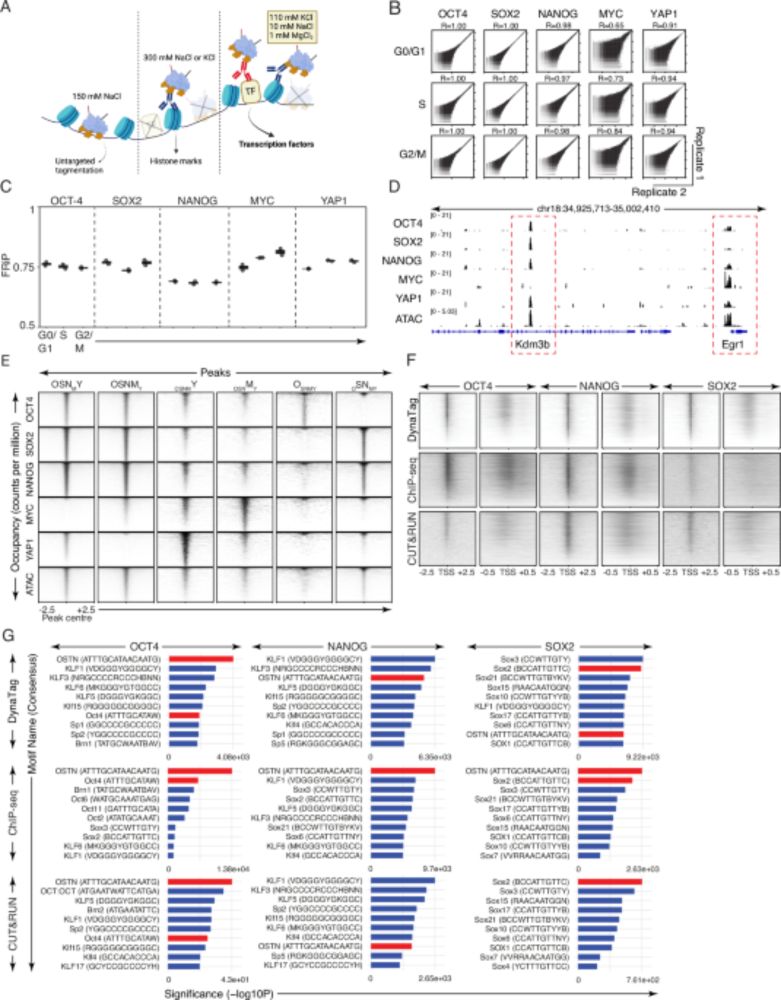
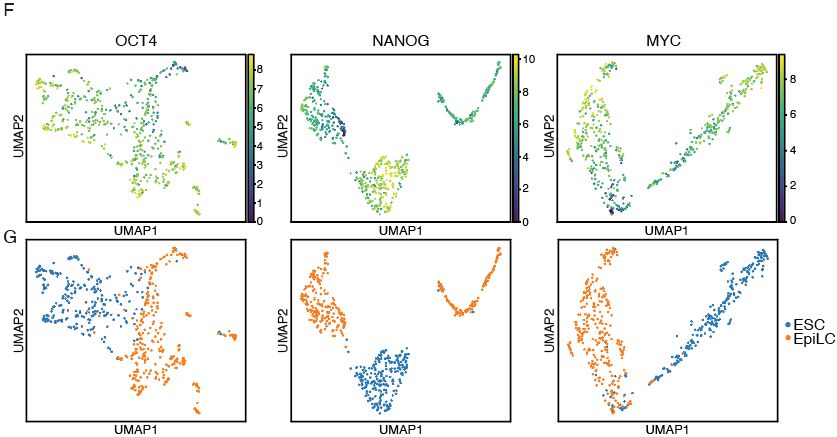
⚡Preserves weak/transient TF binding without signal loss
⚡Better signal-to-noise & resolution
⚡Works for all TFs (high and low DNA-binding affinity) & histone marks
⚡Low-input needed, bulk or single-cell, scalable to multi-cellular systems (4/7)
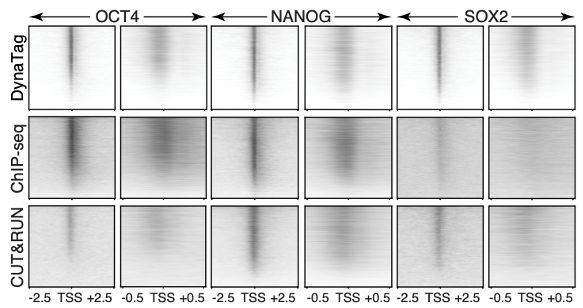
⚡Preserves weak/transient TF binding without signal loss
⚡Better signal-to-noise & resolution
⚡Works for all TFs (high and low DNA-binding affinity) & histone marks
⚡Low-input needed, bulk or single-cell, scalable to multi-cellular systems (4/7)
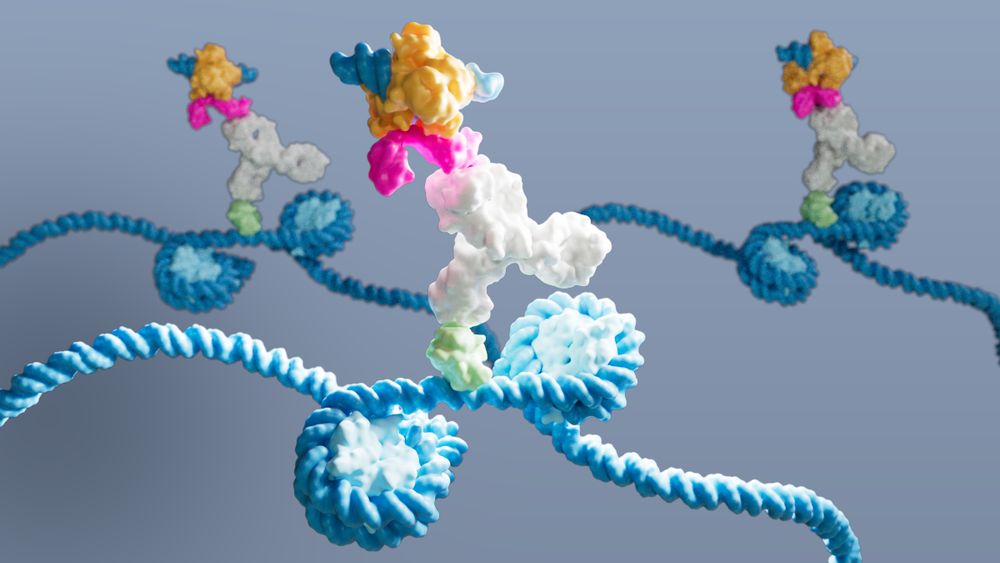
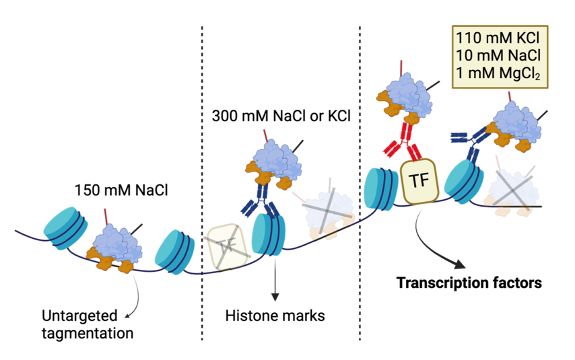
💡 Enter #DynaTag: ‘Dynamic targets and Tagmentation’ captures transient TF-DNA interactions that standard CUT&Tag misses.
The trick? Sample prep under physiological salt stabilizes specific interactions without promoting untargeted tagmentation 🧂 (3/7)
💡 Enter #DynaTag: ‘Dynamic targets and Tagmentation’ captures transient TF-DNA interactions that standard CUT&Tag misses.
The trick? Sample prep under physiological salt stabilizes specific interactions without promoting untargeted tagmentation 🧂 (3/7)
👉 Doesn’t work well for TFs that only transiently bind DNA
👉 These fleeting interactions are often lost during sample prep
That means we’re missing key regulatory events and it’s time for a new method (2/7)
👉 Doesn’t work well for TFs that only transiently bind DNA
👉 These fleeting interactions are often lost during sample prep
That means we’re missing key regulatory events and it’s time for a new method (2/7)

