
I engineer and optimize the metabolism of nonmodel C1-trophic bacteria for novel biotechnological applications
Metabolic Engineering | Green Economy | CO2 valorization | DTU Biosustain
👉 researchleaderprogramme.com/recipients/e...

👉 researchleaderprogramme.com/recipients/e...
With a turnaround time that is 50% shorter than the current systems available, we believe that SIBR-Cas9 and SIBR2.0-Cas12a will help using C. necator in new P2X applications! 8/n

With a turnaround time that is 50% shorter than the current systems available, we believe that SIBR-Cas9 and SIBR2.0-Cas12a will help using C. necator in new P2X applications! 8/n



Targeting worked fine & by combining SIBR w/ Cas9, we achieved high editing efficiencies of >80% in two loci! Great start! 4/n

Targeting worked fine & by combining SIBR w/ Cas9, we achieved high editing efficiencies of >80% in two loci! Great start! 4/n





www.nature.com/articles/s41...
www.nature.com/articles/s41...





doi.org/10.1101/2024...

doi.org/10.1101/2024...
doi.org/10.1016/j.co...

doi.org/10.1016/j.co...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

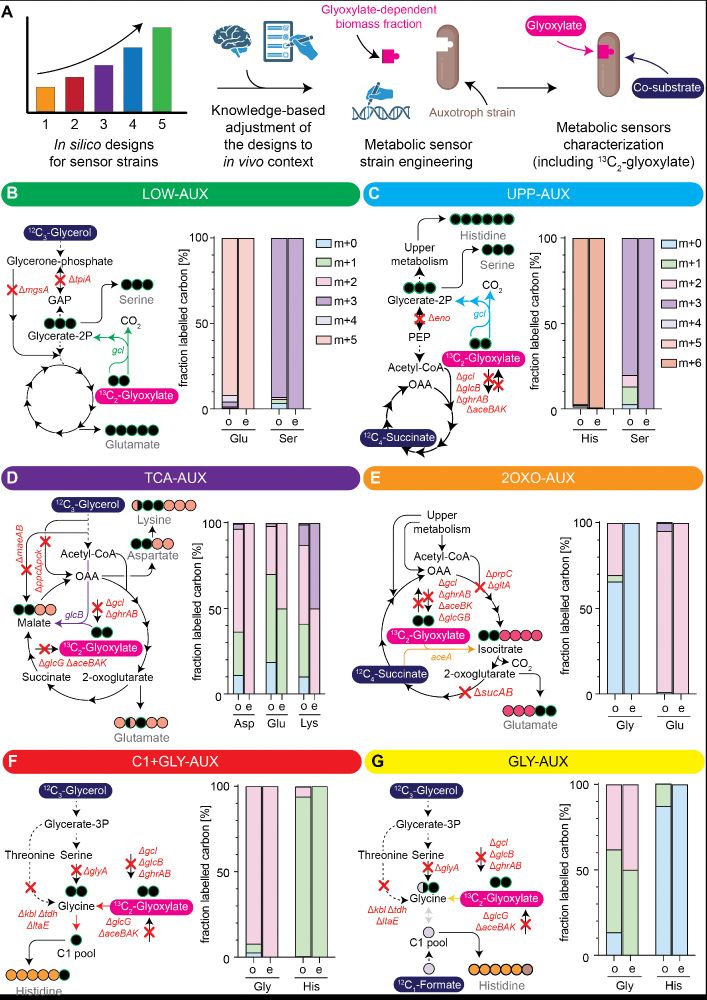
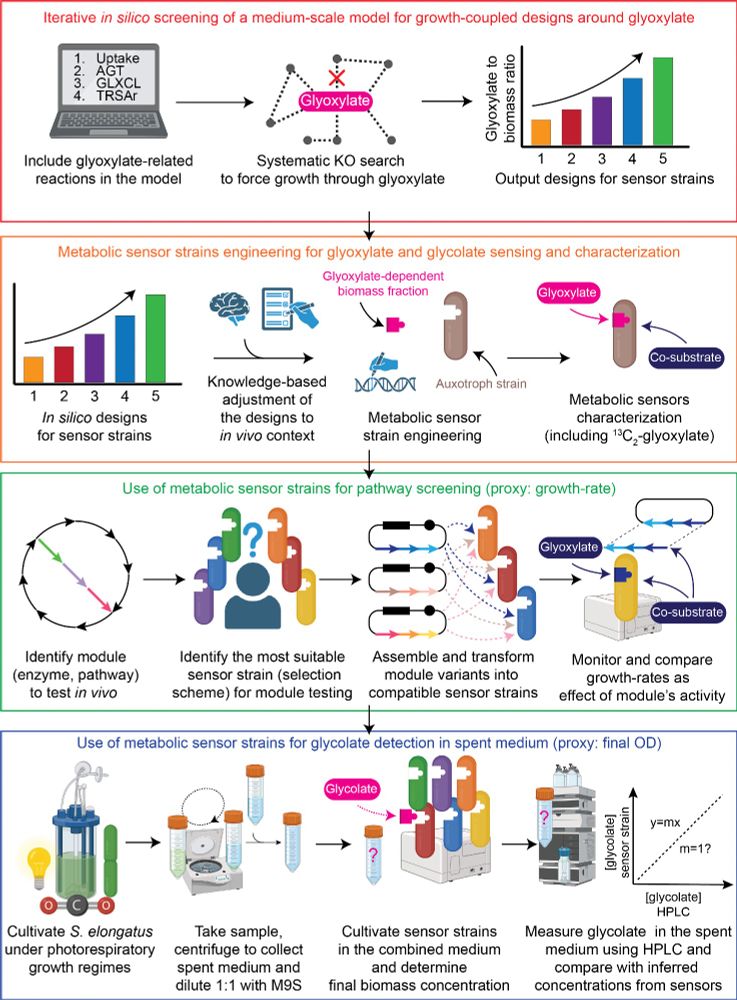
To implement these solutions, we can create auxotrophic metabolic sensors, coupling growth to target metabolic module activity. In this #preprint co-led with Helena Schulz, we develop a suite of glyoxylate and glycolate sensors with wide-range sensitivity & different scopes
doi.org/10.1016/j.co...

To implement these solutions, we can create auxotrophic metabolic sensors, coupling growth to target metabolic module activity. In this #preprint co-led with Helena Schulz, we develop a suite of glyoxylate and glycolate sensors with wide-range sensitivity & different scopes
doi.org/10.1016/j.co...
doi.org/10.1016/j.co...

doi.org/10.1016/j.co...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
The system is based on self-splicing introns and allows us to get mutants within 48h and with efficiencies up to 80%
doi.org/10.1101/2024.11.24.625072 [1/n]

The system is based on self-splicing introns and allows us to get mutants within 48h and with efficiencies up to 80%
doi.org/10.1101/2024.11.24.625072 [1/n]

