
CRUK-SI: https://tinyurl.com/5fr9n2ew
QUB: https://tinyurl.com/4b68vf56
Excellent presention from Raheleh in our team charting phenotypic evolution during cancer development and progression, and how best we can use preclinical models of cancer to improve patient outcomes


Excellent presention from Raheleh in our team charting phenotypic evolution during cancer development and progression, and how best we can use preclinical models of cancer to improve patient outcomes
#AACR25


#AACR25
Understanding precancer biology is crucial for improving early detection, and preventing early cancer development and progression.
#AACR25
@nataliefisher.bsky.social


Understanding precancer biology is crucial for improving early detection, and preventing early cancer development and progression.
#AACR25
@nataliefisher.bsky.social

Natalie's @theaacr.bsky.social Women in Cancer Research award talk covers how we can use biology to improve early detection of polyps
Sid's poster is on early tumour transcriptional landscapes


Natalie's @theaacr.bsky.social Women in Cancer Research award talk covers how we can use biology to improve early detection of polyps
Sid's poster is on early tumour transcriptional landscapes


(no one on bluesky yet!)

(no one on bluesky yet!)
This patient group has the highest risk of disease relapse and unfortunately does not response to current 5FU-based therapies
So a BIG opportunity here

This patient group has the highest risk of disease relapse and unfortunately does not response to current 5FU-based therapies
So a BIG opportunity here
and using polyI:C we could push cancer stem cells between these states!!


and using polyI:C we could push cancer stem cells between these states!!

but what was most interesting was the contrasting phenotypes this signalling induced prior to the anti-metastatic CD8+ liver response we see at the end

but what was most interesting was the contrasting phenotypes this signalling induced prior to the anti-metastatic CD8+ liver response we see at the end
So characterised a panel of these lineages
& see that macrophages love it, but (some) cancer cells not so much


So characterised a panel of these lineages
& see that macrophages love it, but (some) cancer cells not so much


which can be therapeutically activated using poly(I:C)..
gut.bmj.com/content/71/12/

which can be therapeutically activated using poly(I:C)..
gut.bmj.com/content/71/12/
TLDR; aggressive CMS4 tumours can be reprogrammed towards less aggressive CMS1
biorxiv.org/content/10.1...
thread 👇
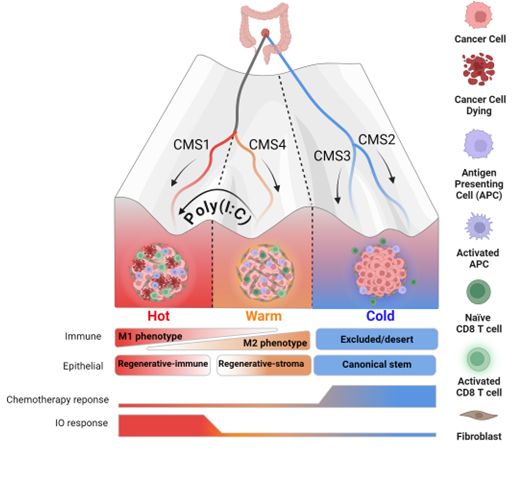
TLDR; aggressive CMS4 tumours can be reprogrammed towards less aggressive CMS1
biorxiv.org/content/10.1...
thread 👇






In addition, these results are always consistent and highly significant regardless of what GSEA method used

In addition, these results are always consistent and highly significant regardless of what GSEA method used



