
My proposal: 'ArmoredPhage' 🛡️
My goal: Engineer a Thermostable MS2 Bacteriophage Using Protein Design Techniques.
Check it out:
djosergenomics.github.io/Armored-Phag...
#HTGAA #synbio #AlphaFold #proteindesign



My proposal: 'ArmoredPhage' 🛡️
My goal: Engineer a Thermostable MS2 Bacteriophage Using Protein Design Techniques.
Check it out:
djosergenomics.github.io/Armored-Phag...
#HTGAA #synbio #AlphaFold #proteindesign
I was always intimidated by 3D protein models in papers & never thought I'd understand them one day!
Today, I loaded a PDB file into PyMOL, saw the alpha helices & beta sheets of my Tyrosinase enzyme.
This is so cool. 🚀
#HTGAA #proteindesign


I was always intimidated by 3D protein models in papers & never thought I'd understand them one day!
Today, I loaded a PDB file into PyMOL, saw the alpha helices & beta sheets of my Tyrosinase enzyme.
This is so cool. 🚀
#HTGAA #proteindesign
I wrote a script to program a virtual Opentrons robot to draw my Djoser Genomics pyramid. It simulates robots pipetting 100+ dots of fluorescent E.coli because manually it is slow & error-prone.
#HTGAA #synbio #Opentrons #automation #bioinformatics

I wrote a script to program a virtual Opentrons robot to draw my Djoser Genomics pyramid. It simulates robots pipetting 100+ dots of fluorescent E.coli because manually it is slow & error-prone.
#HTGAA #synbio #Opentrons #automation #bioinformatics
The Grand Finale, we dive into Gene set Enrichment Analysis (GSEA), why we do it, how we do it and how to visualize and interpret it.
Check it out: djosergenomics.github.io/Scroll-8-Djo...
#bioinformatics #RNAseq #scicomm

The Grand Finale, we dive into Gene set Enrichment Analysis (GSEA), why we do it, how we do it and how to visualize and interpret it.
Check it out: djosergenomics.github.io/Scroll-8-Djo...
#bioinformatics #RNAseq #scicomm
Here, We identify Trends among our DEGs using Functional (GO) and Pathway (KEGG) Enrichment Analysis!
Check it out: djosergenomics.github.io/Scroll-7-Thu...
#bioinformatics #RNAseq #scicomm

Here, We identify Trends among our DEGs using Functional (GO) and Pathway (KEGG) Enrichment Analysis!
Check it out: djosergenomics.github.io/Scroll-7-Thu...
#bioinformatics #RNAseq #scicomm
We dive into Visualizations!
MA, PCA, Volcano Plots and Heatmaps!
Check it out: djosergenomics.github.io/Scroll-6-Ram...
#bioinformatics #RNAseq #scicomm

We dive into Visualizations!
MA, PCA, Volcano Plots and Heatmaps!
Check it out: djosergenomics.github.io/Scroll-6-Ram...
#bioinformatics #RNAseq #scicomm
We dive into Differential expression analysis (DEA), using DESeq2, and how to interpret the findings
Check it out: djosergenomics.github.io/Scroll-5-Men...
#bioinformatics #RNAseq #scicomm

We dive into Differential expression analysis (DEA), using DESeq2, and how to interpret the findings
Check it out: djosergenomics.github.io/Scroll-5-Men...
#bioinformatics #RNAseq #scicomm
We step into the next stage of our RNAseq journey!
Importing Kallisto results into R, linking them to gene annotations, & building a strong foundation for downstream analysis
Here: djosergenomics.github.io/Scroll-4-Kha...
#bioinformatics #RNAseq #scicomm

We step into the next stage of our RNAseq journey!
Importing Kallisto results into R, linking them to gene annotations, & building a strong foundation for downstream analysis
Here: djosergenomics.github.io/Scroll-4-Kha...
#bioinformatics #RNAseq #scicomm
⚒️ Guided by King Khufu, we pseudoalign our reads using Kallisto — fast, efficient, and accurate.
🌐 Full tutorial + cultural spotlight:
📜 djosergenomics.github.io/Scroll-3-Khu...
#RNAseq #bioinformatics #scicomm

⚒️ Guided by King Khufu, we pseudoalign our reads using Kallisto — fast, efficient, and accurate.
🌐 Full tutorial + cultural spotlight:
📜 djosergenomics.github.io/Scroll-3-Khu...
#RNAseq #bioinformatics #scicomm
We dive into RNA-seq Quality Control with FASTQC — guided by Hesy-Ra, the first known dentist & physician in history.
🧪 Read here: djosergenomics.github.io/Scroll-2-Hes...
#bioinformatics #RNAseq #scicomm

We dive into RNA-seq Quality Control with FASTQC — guided by Hesy-Ra, the first known dentist & physician in history.
🧪 Read here: djosergenomics.github.io/Scroll-2-Hes...
#bioinformatics #RNAseq #scicomm
We start by collecting real RNA-seq data from ENA using Google Colab.
Inspired by Imhotep - ancient Egypt’s architect of the Pyramid of Djoser.
📜 Check it out: djosergenomics.github.io/Scroll-1-Imh...
#bioinformatics #RNAseq

We start by collecting real RNA-seq data from ENA using Google Colab.
Inspired by Imhotep - ancient Egypt’s architect of the Pyramid of Djoser.
📜 Check it out: djosergenomics.github.io/Scroll-1-Imh...
#bioinformatics #RNAseq
📊 40 significant pathways (padj < 0.05, |NES| > 1) highlighting strong immune and inflammatory signals.
🏆 Top enriched pathway: KEGG Allograft Rejection
(NES = 2.44, p.adj = 1.87e-3)
Suggests a major immune activation signature 🚨
#Bioinformatics #RNAseq #GSEA

📊 40 significant pathways (padj < 0.05, |NES| > 1) highlighting strong immune and inflammatory signals.
🏆 Top enriched pathway: KEGG Allograft Rejection
(NES = 2.44, p.adj = 1.87e-3)
Suggests a major immune activation signature 🚨
#Bioinformatics #RNAseq #GSEA
🧪 Top 10 significant enriched pathways include:
🔬 Infectious disease pathways (COVID-19, Tuberculosis, Staphylococcus aureus)
🔬 Ribosomal activity & phagosome function
🔬 Immune-driven diseases (Asthma, Alcoholic liver disease)
#Bioinformatics #RNAseq #KEGG

🧪 Top 10 significant enriched pathways include:
🔬 Infectious disease pathways (COVID-19, Tuberculosis, Staphylococcus aureus)
🔬 Ribosomal activity & phagosome function
🔬 Immune-driven diseases (Asthma, Alcoholic liver disease)
#Bioinformatics #RNAseq #KEGG
Functional enrichment analysis reveals 148 significant GO terms (padj < 0.05), clustering into key biological processes:
💡 BP: Immune response, Cell Adhesion & Complement Activation
💡 CC & MF: Ribosomes & MHC class I & II Protein Complexes
#Bioinformatics #RNAseq #GO

Functional enrichment analysis reveals 148 significant GO terms (padj < 0.05), clustering into key biological processes:
💡 BP: Immune response, Cell Adhesion & Complement Activation
💡 CC & MF: Ribosomes & MHC class I & II Protein Complexes
#Bioinformatics #RNAseq #GO
📌 HS02 & CD01 are the most different.
📌 HS02 & HS03 cluster together, suggesting shared expression.
📌 CD03 appears distinct from other CD samples, possibly due to biological or technical factors.
#RNAseq #Bioinformatics

📌 HS02 & CD01 are the most different.
📌 HS02 & HS03 cluster together, suggesting shared expression.
📌 CD03 appears distinct from other CD samples, possibly due to biological or technical factors.
#RNAseq #Bioinformatics
📊 HS02 & HS03 have a slight outlier.
📊 CD03 follows expected trends.
#Bioinformatics #RNAseq

📊 HS02 & HS03 have a slight outlier.
📊 CD03 follows expected trends.
#Bioinformatics #RNAseq
🧬 Most genes stay around log2FC = 0, while some show strong up/downregulation.
🧬 These genes are potential candidates for further functional analysis.
#Bioinformatics #RNAseq

🧬 Most genes stay around log2FC = 0, while some show strong up/downregulation.
🧬 These genes are potential candidates for further functional analysis.
#Bioinformatics #RNAseq
🟥🟦 The grouping indicates consistent transcriptomic differences between healthy and diseased conditions.
#Bioinformatics #RNAseq #Transcriptomics

🟥🟦 The grouping indicates consistent transcriptomic differences between healthy and diseased conditions.
#Bioinformatics #RNAseq #Transcriptomics
🔥 It shows differentially expressed genes with log2FC > 1 or < -1 & padj < 0.05 are in red.
🔥 These genes are the most significantly dysregulated and could be important in disease mechanisms.
#Bioinformatics #RNAseq

🔥 It shows differentially expressed genes with log2FC > 1 or < -1 & padj < 0.05 are in red.
🔥 These genes are the most significantly dysregulated and could be important in disease mechanisms.
#Bioinformatics #RNAseq
🔹 PC1 explains 39.2% of variance, PC2 26.7%.
I realized I mistakenly swapped the Disease and Healthy samples in my old PCA plot. 😅🙈
#Bioinformatics #RNAseq
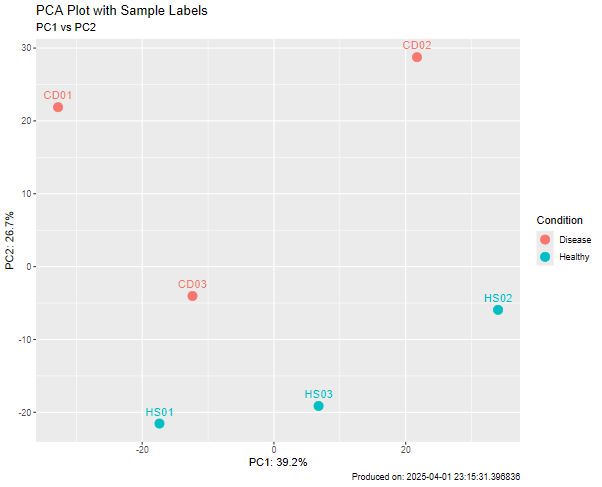
🔹 PC1 explains 39.2% of variance, PC2 26.7%.
I realized I mistakenly swapped the Disease and Healthy samples in my old PCA plot. 😅🙈
#Bioinformatics #RNAseq
📌 CD01 & CD02 are highly similar.
📌 HS02 & CD03 are the most different.
📌 HS01 & HS02 cluster together, suggesting shared expression.
📌 CD03 appears distinct from other CD samples, possibly due to biological or technical factors.
#RNAseq #Bioinformatics

📌 CD01 & CD02 are highly similar.
📌 HS02 & CD03 are the most different.
📌 HS01 & HS02 cluster together, suggesting shared expression.
📌 CD03 appears distinct from other CD samples, possibly due to biological or technical factors.
#RNAseq #Bioinformatics
📊 HS02 has a slight outlier.
📊 CD01 follows expected trends.
#Bioinformatics #RNAseq

📊 HS02 has a slight outlier.
📊 CD01 follows expected trends.
#Bioinformatics #RNAseq
🧬 Most genes stay around log2FC = 0, while some show strong up/downregulation.
🧬 These genes are potential candidates for further functional analysis.
#Bioinformatics #RNAseq

🧬 Most genes stay around log2FC = 0, while some show strong up/downregulation.
🧬 These genes are potential candidates for further functional analysis.
#Bioinformatics #RNAseq
🟥🟦 The grouping indicates consistent transcriptomic differences between healthy and diseased conditions.
#Bioinformatics #RNAseq #Transcriptomics

🟥🟦 The grouping indicates consistent transcriptomic differences between healthy and diseased conditions.
#Bioinformatics #RNAseq #Transcriptomics
🔥 It shows differentially expressed genes with log2FC > 1 or < -1 & padj < 0.05 are in red.
🔥 These genes are the most significantly dysregulated and could be important in disease mechanisms.
#Bioinformatics #RNAseq

🔥 It shows differentially expressed genes with log2FC > 1 or < -1 & padj < 0.05 are in red.
🔥 These genes are the most significantly dysregulated and could be important in disease mechanisms.
#Bioinformatics #RNAseq

