Elzo de Wit lab @ NKI
@dewitlab.bsky.social
Computational & Genomics Lab studying 3D genome function and dynamics @ Netherlands Cancer Institute
Very interesting indeed! Also wondering what we see here are extrusion fountains in the RBCs?

August 26, 2025 at 12:22 PM
Very interesting indeed! Also wondering what we see here are extrusion fountains in the RBCs?
At the level of TADs we also saw intriguing differences. Below you can see an example of a region, where in one of the patient samples the TAD structure seems to be almost completely gone. We currently do no understand what causes these differences in TAD strength.

August 18, 2025 at 2:41 PM
At the level of TADs we also saw intriguing differences. Below you can see an example of a region, where in one of the patient samples the TAD structure seems to be almost completely gone. We currently do no understand what causes these differences in TAD strength.
However(!), when we looked at late stage cancer biopsies that were acquired from pleural effusions, we observed massive differences between patients. Both compartment strength and compartment identity was highly variable between patients.

August 18, 2025 at 2:41 PM
However(!), when we looked at late stage cancer biopsies that were acquired from pleural effusions, we observed massive differences between patients. Both compartment strength and compartment identity was highly variable between patients.
When we compared the 3D genome of primary tumors to those of metastatic samples we were once again surprised by the similarity. We also had difficulty identifying consistent changes among all the patients.

August 18, 2025 at 2:41 PM
When we compared the 3D genome of primary tumors to those of metastatic samples we were once again surprised by the similarity. We also had difficulty identifying consistent changes among all the patients.
Also at the level of compartments there seems to be limited differences in compartment scores and there do not seem to be outright switches of compartments from A to B or vice versa when we compared the primary tumor to the healthy tissue.

August 18, 2025 at 2:41 PM
Also at the level of compartments there seems to be limited differences in compartment scores and there do not seem to be outright switches of compartments from A to B or vice versa when we compared the primary tumor to the healthy tissue.
We performed in situ Hi-C on the samples to determine the 3D genome organization.
To our surprise, at the assayed resolution, the Hi-C maps look quite similar between healthy breast and primary tumor. Below this can be seen for the insulation score, which indicates the position of TAD boundaries.
To our surprise, at the assayed resolution, the Hi-C maps look quite similar between healthy breast and primary tumor. Below this can be seen for the insulation score, which indicates the position of TAD boundaries.

August 18, 2025 at 2:41 PM
We performed in situ Hi-C on the samples to determine the 3D genome organization.
To our surprise, at the assayed resolution, the Hi-C maps look quite similar between healthy breast and primary tumor. Below this can be seen for the insulation score, which indicates the position of TAD boundaries.
To our surprise, at the assayed resolution, the Hi-C maps look quite similar between healthy breast and primary tumor. Below this can be seen for the insulation score, which indicates the position of TAD boundaries.
Strong The Onion vibes with this one: www.nature.com/articles/d41...

July 16, 2025 at 8:13 PM
Strong The Onion vibes with this one: www.nature.com/articles/d41...
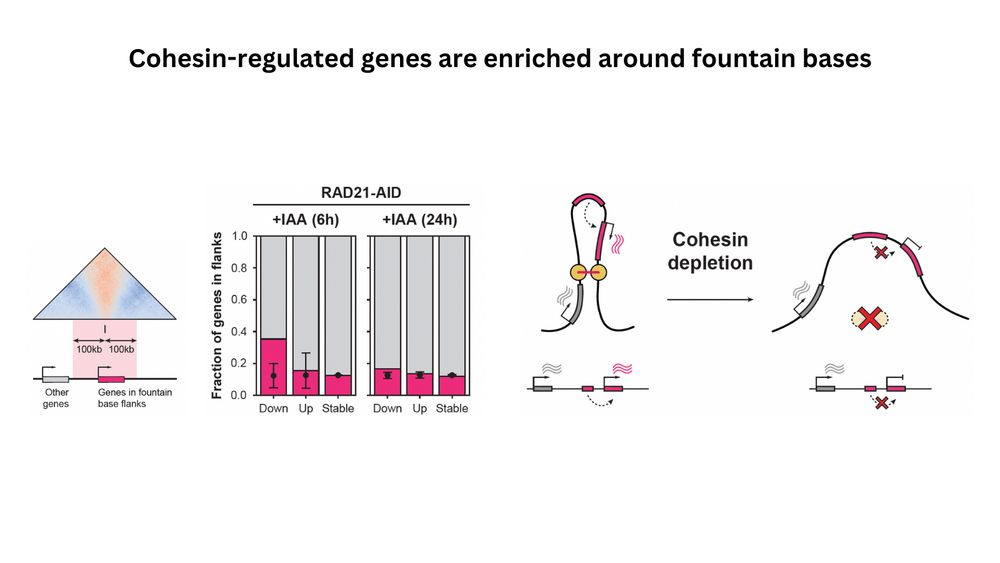
(8/n) Do fountains have a functional role? Our RNA-seq data revealed that their disruption resulted in downregulation of genes around fountains. This would be consistent with a model where cohesin loading at enhancers promotes enhancer-promoter communication and activates genes.
July 1, 2025 at 6:34 AM
(8/n) Do fountains have a functional role? Our RNA-seq data revealed that their disruption resulted in downregulation of genes around fountains. This would be consistent with a model where cohesin loading at enhancers promotes enhancer-promoter communication and activates genes.
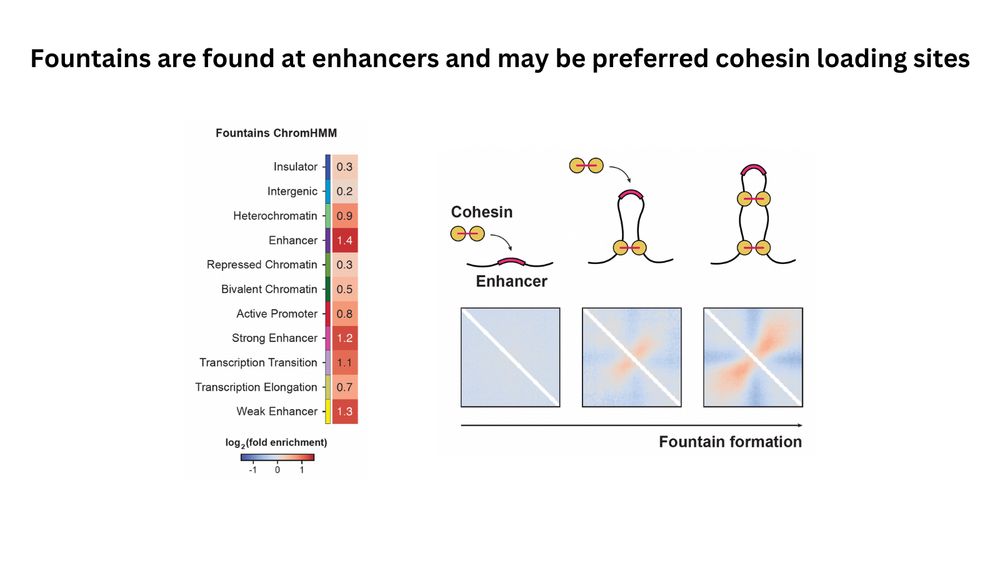
(7/n) Where do fountains originate from? We found that fountains are enriched in enhancer chromatin features, consistent with observations in other species that fountains are formed at regions of active chromatin.
July 1, 2025 at 6:34 AM
(7/n) Where do fountains originate from? We found that fountains are enriched in enhancer chromatin features, consistent with observations in other species that fountains are formed at regions of active chromatin.
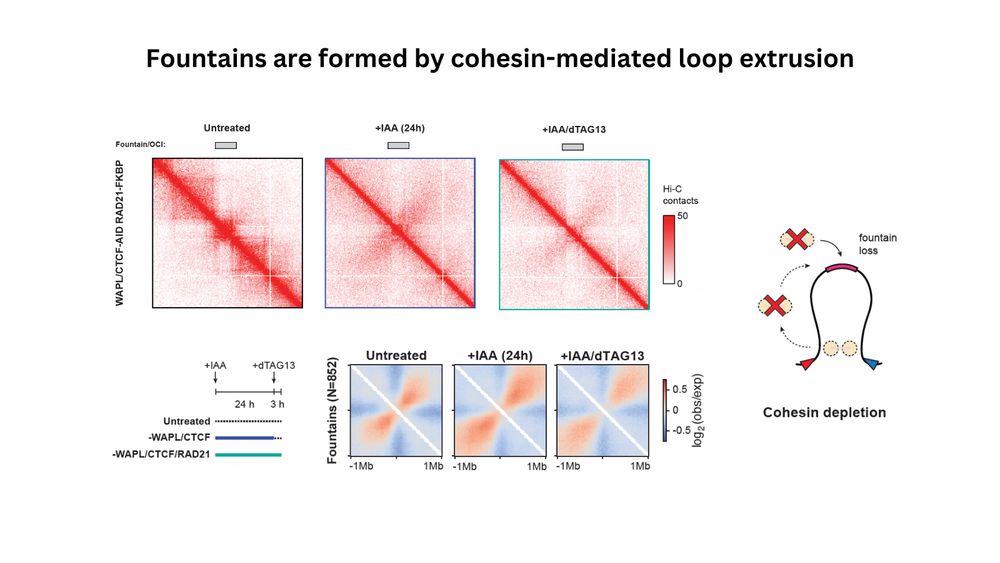
(5/n) Are fountains then a consequence of loop extrusion? To understand this, we tagged RAD21 in the WAPL/CTCF background, making it a triple degron cell line. Sequential depletion experiments clearly demonstrated that fountains are dependent on cohesin and reduced in its absence.
July 1, 2025 at 6:34 AM
(5/n) Are fountains then a consequence of loop extrusion? To understand this, we tagged RAD21 in the WAPL/CTCF background, making it a triple degron cell line. Sequential depletion experiments clearly demonstrated that fountains are dependent on cohesin and reduced in its absence.
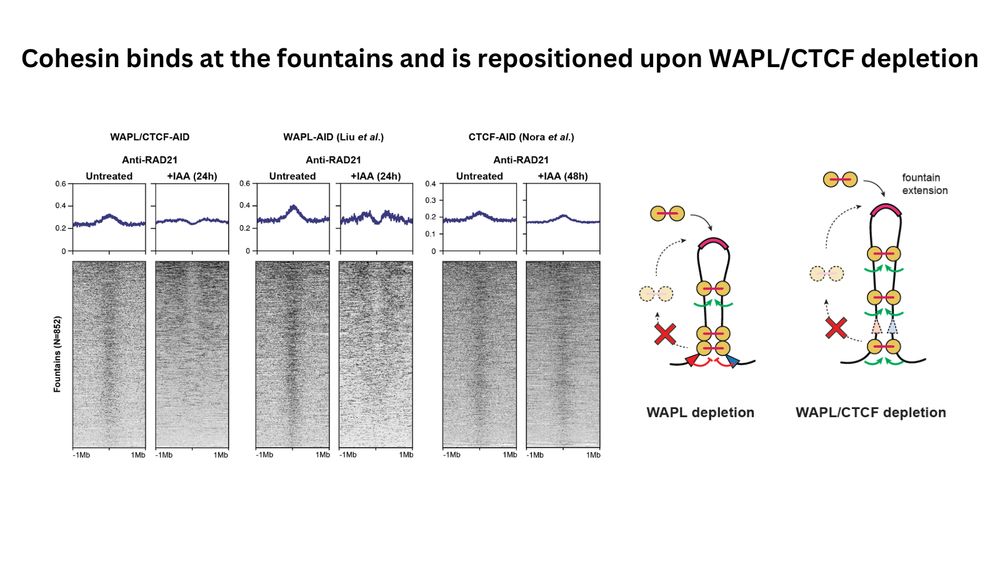
(4/n) What is the mechanism behind their formation? Regions making up the fountains exhibit strong cohesin binding, which is repositioned away upon WAPL and CTCF depletion. Additional analyses of WAPL and CTCF single depletions, suggests that cohesin might be loaded directly at the fountain base.
July 1, 2025 at 6:34 AM
(4/n) What is the mechanism behind their formation? Regions making up the fountains exhibit strong cohesin binding, which is repositioned away upon WAPL and CTCF depletion. Additional analyses of WAPL and CTCF single depletions, suggests that cohesin might be loaded directly at the fountain base.
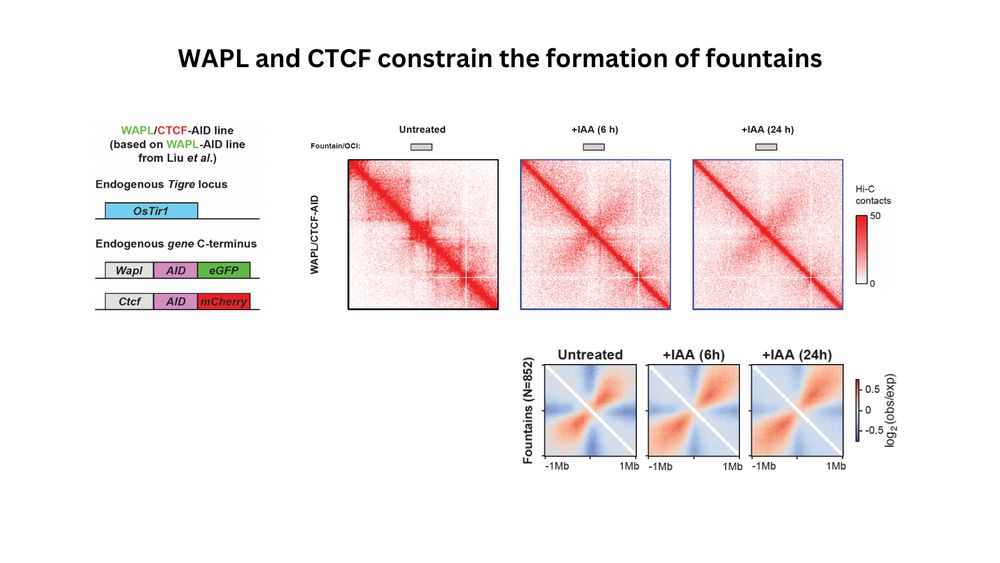
(3/n) Systematic analysis of fountains revealed that they can already be visible as a fountain-like pattern in the unperturbed cells. Fountains are extended upon depletion of WAPL and CTCF, which constrain fountain dynamics.
July 1, 2025 at 6:34 AM
(3/n) Systematic analysis of fountains revealed that they can already be visible as a fountain-like pattern in the unperturbed cells. Fountains are extended upon depletion of WAPL and CTCF, which constrain fountain dynamics.
In an in vitro embryo model (‘gastruloids’), we find that loss of ZFP143 reduces growth and severely hampers development. A decrease in mitochondrial membrane potential suggests that also in gastruloids mitochondrial function is disrupted.

December 20, 2024 at 4:22 PM
In an in vitro embryo model (‘gastruloids’), we find that loss of ZFP143 reduces growth and severely hampers development. A decrease in mitochondrial membrane potential suggests that also in gastruloids mitochondrial function is disrupted.
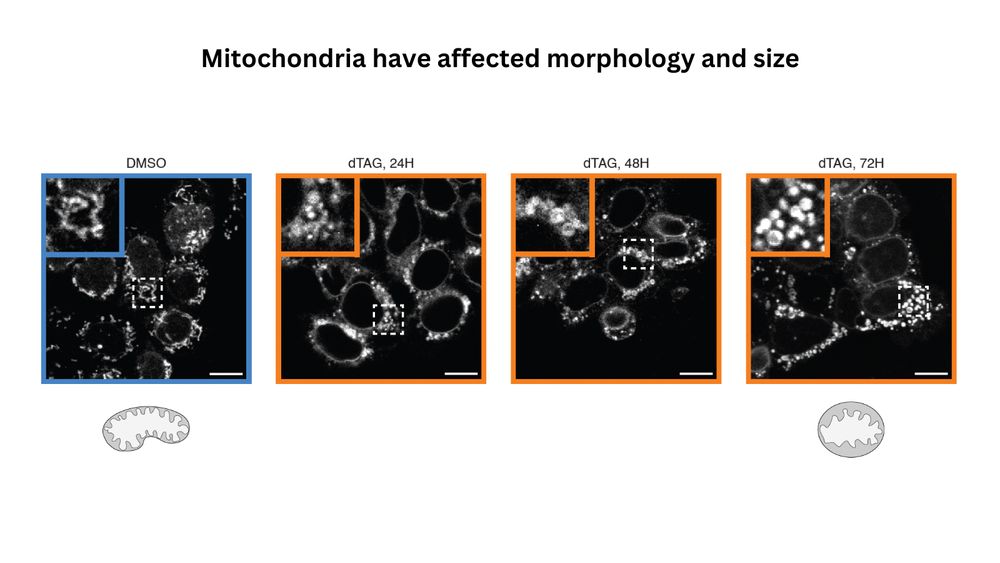
To check this we performed TT-seq and RNAseq following depletion of ZFP143. We find that many nuclear encoded mitochondrial genes are downregulated in the absence of ZFP143. This downregulation also affected mitochondrial morphology and proliferation.
December 20, 2024 at 4:22 PM
To check this we performed TT-seq and RNAseq following depletion of ZFP143. We find that many nuclear encoded mitochondrial genes are downregulated in the absence of ZFP143. This downregulation also affected mitochondrial morphology and proliferation.
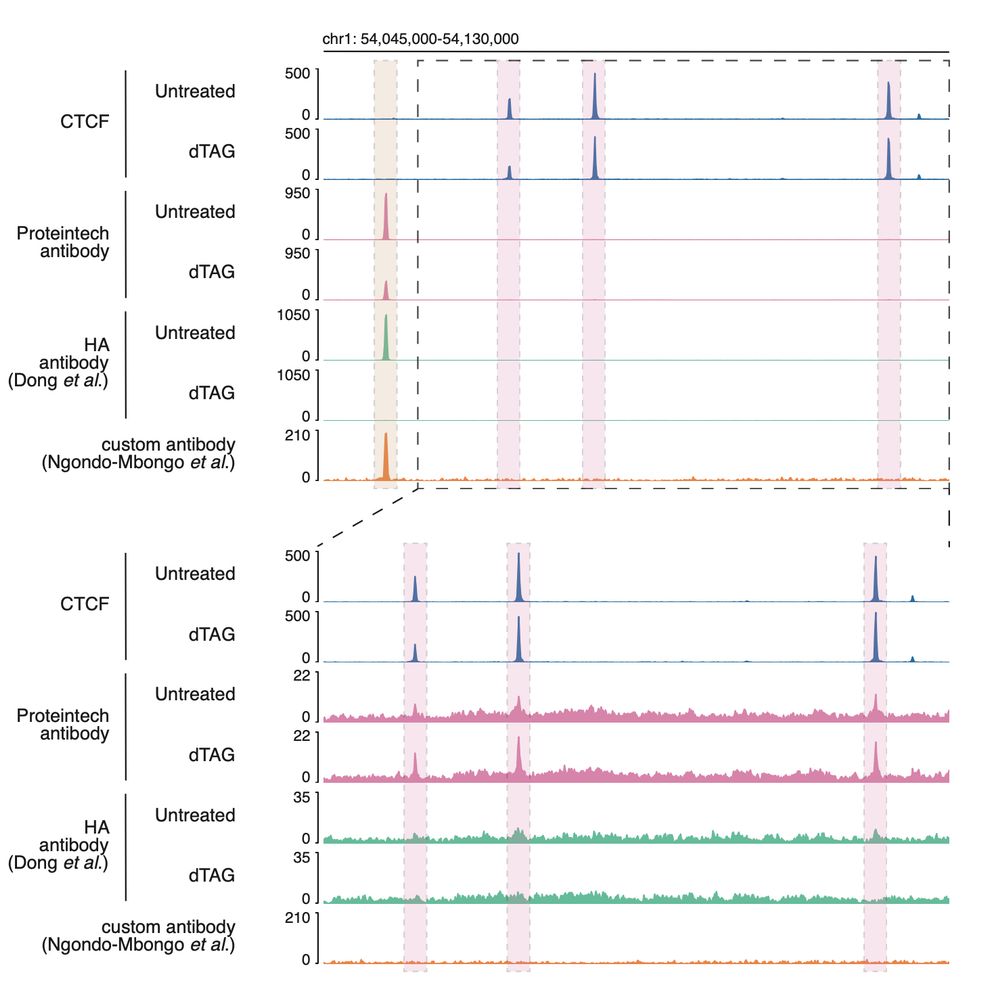
While the “real” ZNF143 peaks are diminished, those overlapping with CTCF binding sites are increased, which is exactly what you would expect in the absence of the primary antibody target.
December 20, 2024 at 4:22 PM
While the “real” ZNF143 peaks are diminished, those overlapping with CTCF binding sites are increased, which is exactly what you would expect in the absence of the primary antibody target.
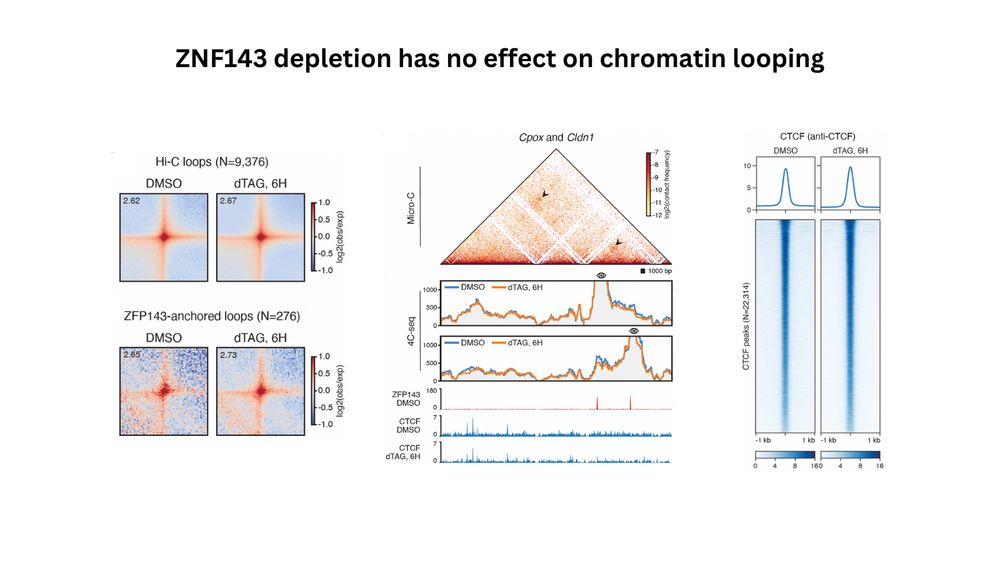
We performed degron experiments for ZFP143 (mouse ZNF143) and could not detect any changes in the Hi-C maps, suggesting that this protein might have nothing to do with looping.
December 20, 2024 at 4:22 PM
We performed degron experiments for ZFP143 (mouse ZNF143) and could not detect any changes in the Hi-C maps, suggesting that this protein might have nothing to do with looping.

