
Previously PhD in Biophysics at the University of Pittsburgh & Carnegie Mellon University.
https://darianyang.github.io
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
We’ve got your back!
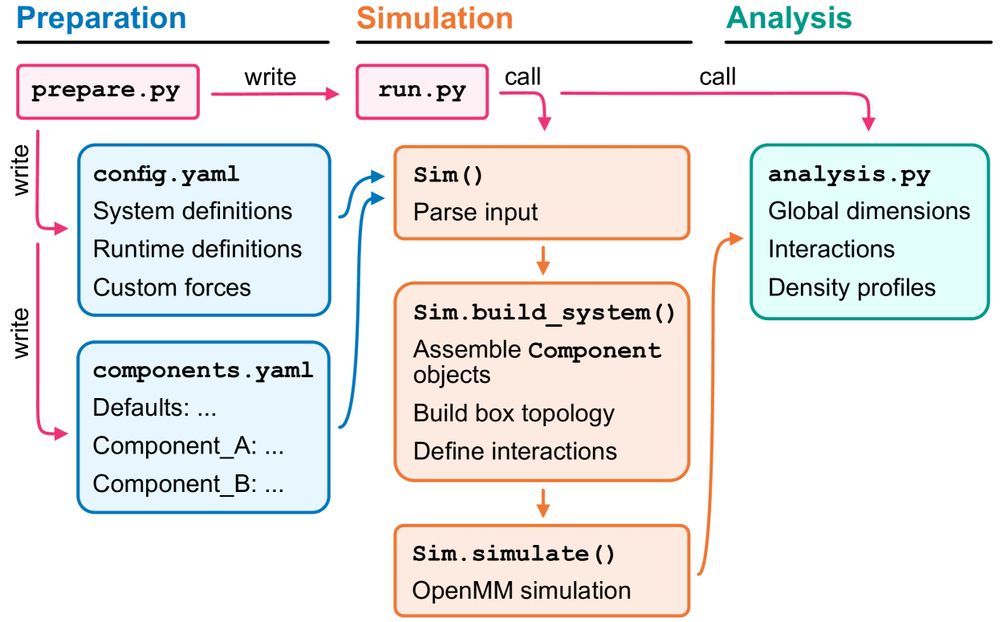
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
📖 Read it here: rdcu.be/ef6YX
📝 Support the statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #collaboration
📖 Read it here: rdcu.be/ef6YX
📝 Support the statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #collaboration
We used 19F NMR and weighted ensemble simulations among other methods to explore hidden dimer states of the HIV-1 capsid protein.
If this sounds interesting to you, see the full paper here:
www.pnas.org/doi/10.1073/...

We used 19F NMR and weighted ensemble simulations among other methods to explore hidden dimer states of the HIV-1 capsid protein.
If this sounds interesting to you, see the full paper here:
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...
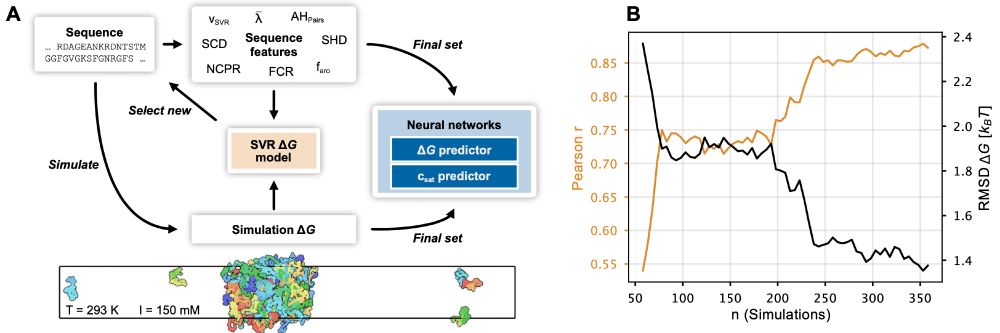
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
📣 NEW BIORXIV ALERT!! 🚨
Using WE MD, linguistic pathway clustering, dynamical network analyses, and HDXMS we reveal a hidden allosteric network within the SARS2 spike S1 domain and predict how the D614G mutation impacts this network!
www.biorxiv.org/content/10.1...

📣 NEW BIORXIV ALERT!! 🚨
Using WE MD, linguistic pathway clustering, dynamical network analyses, and HDXMS we reveal a hidden allosteric network within the SARS2 spike S1 domain and predict how the D614G mutation impacts this network!
www.biorxiv.org/content/10.1...
doi.org/10.1021/acs....
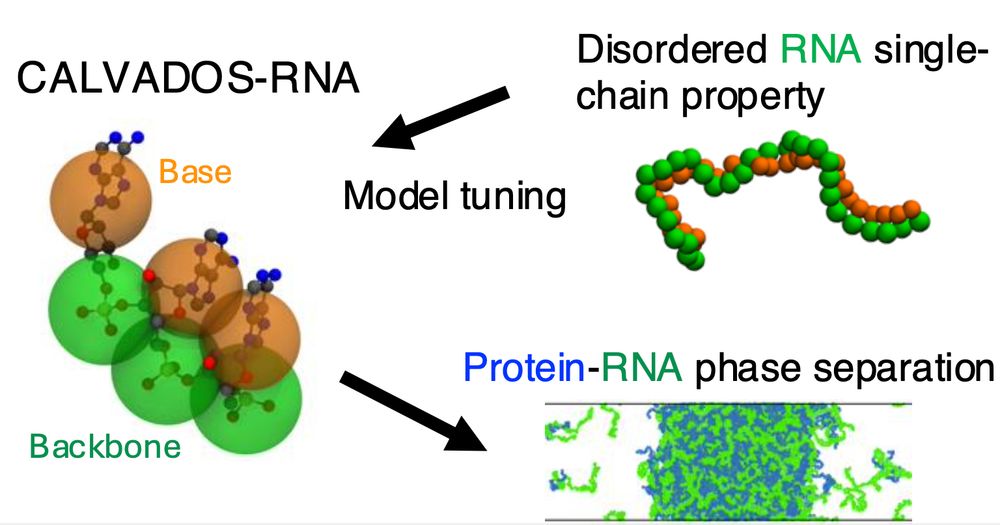
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
doi.org/10.1021/acs....
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
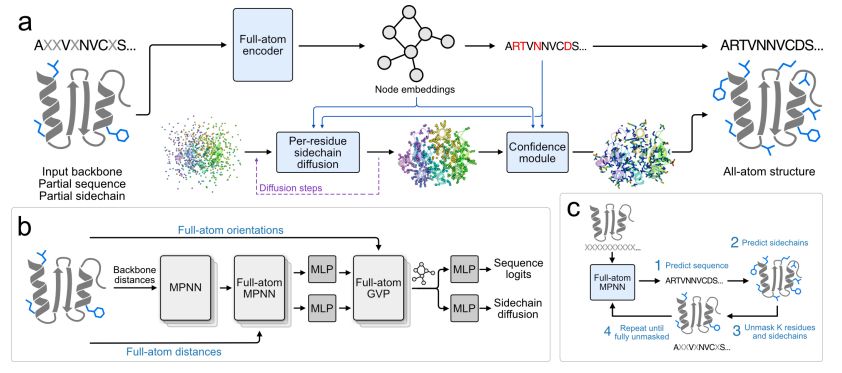
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
🧵👇

www.biorxiv.org/content/10.1...
🧵👇
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
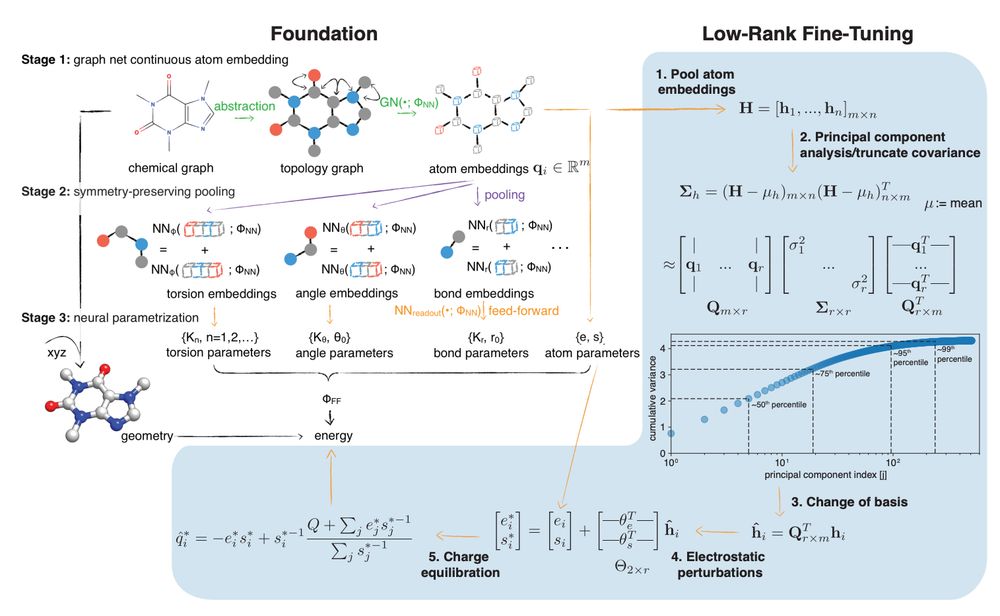
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
We reveal the dynamics and mechanism of target DNA traversal in #CRISPR Cas12a, a conundrum in the field!
nature.com/articles/s41...
#compchem
We thank the amazing #HPC resources of PSC #Anton2 and SDSC
We reveal the dynamics and mechanism of target DNA traversal in #CRISPR Cas12a, a conundrum in the field!
nature.com/articles/s41...
#compchem
We thank the amazing #HPC resources of PSC #Anton2 and SDSC
www.biorxiv.org/content/10.1...
🧵👇 (1/n)

www.biorxiv.org/content/10.1...
🧵👇 (1/n)

@possuhuanglab.bsky.social
www.biorxiv.org/content/10.1...
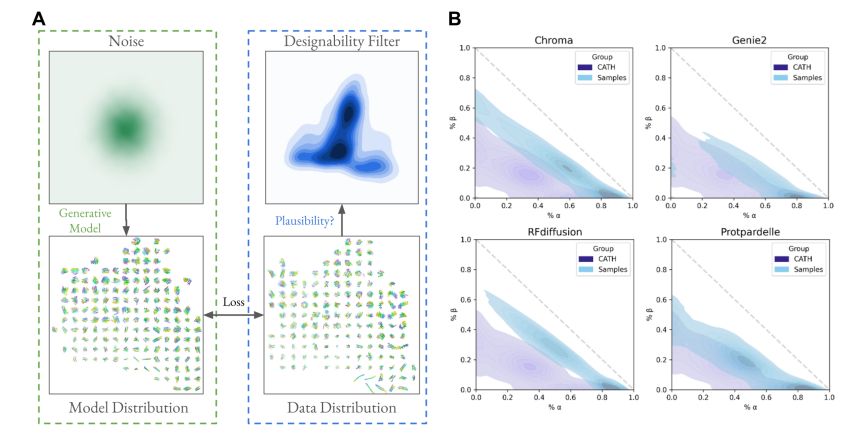
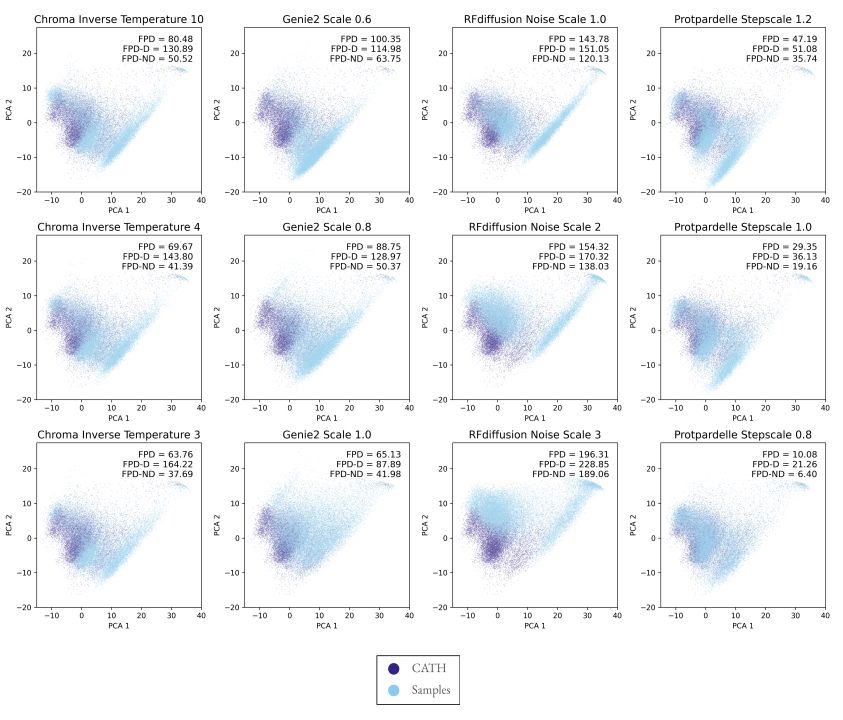
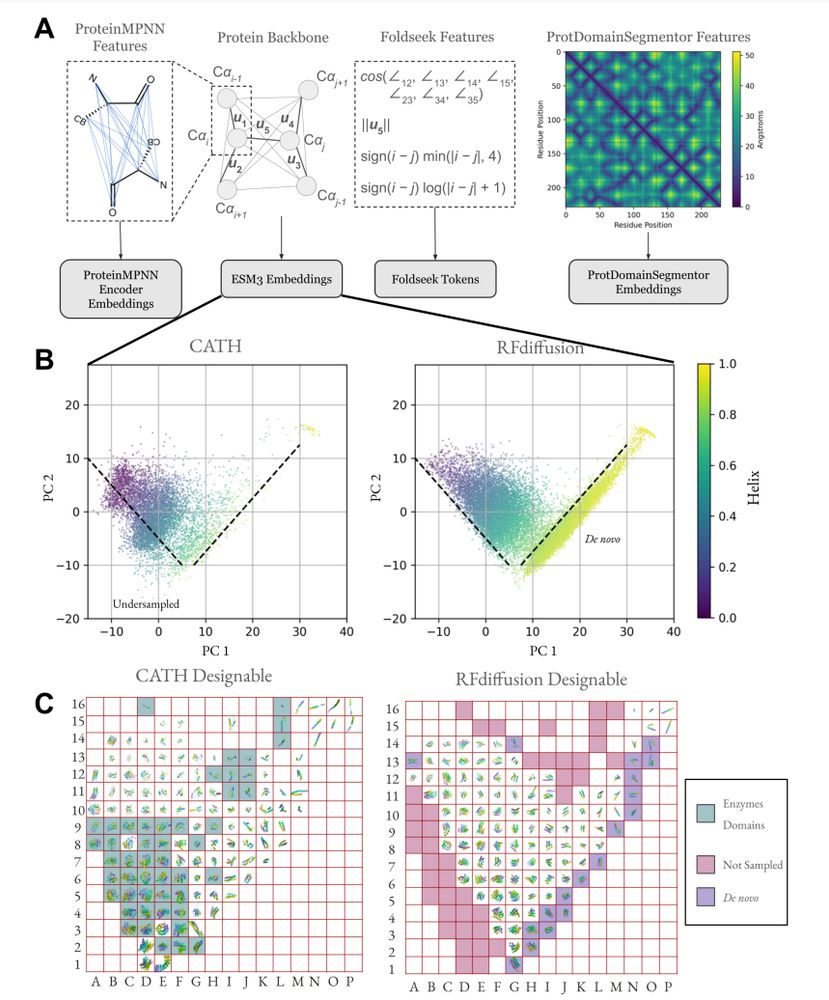
@possuhuanglab.bsky.social
www.biorxiv.org/content/10.1...
Paper led by Jacob Aunstrup from Alex Büll’s lab with MD simulations by Abigail Barclay, and key contributions from several others. We combined measurements of Φ-values with MD simulations to study the transition state for amyloid fibril growth
doi.org/10.1038/s415...

Paper led by Jacob Aunstrup from Alex Büll’s lab with MD simulations by Abigail Barclay, and key contributions from several others. We combined measurements of Φ-values with MD simulations to study the transition state for amyloid fibril growth
doi.org/10.1038/s415...
I'm proud to share our latest study published in hashtag#Nature, driven by Susana Vazquez Torres, and co-led by David Baker (Institute for Protein Design, University of Washington) and myself.


I'm proud to share our latest study published in hashtag#Nature, driven by Susana Vazquez Torres, and co-led by David Baker (Institute for Protein Design, University of Washington) and myself.
if you're interested in the evaluation of conformational accuracy of structure prediction methods, take a look at our first stab at a systematic conformational benchmark in the NP3 technical report below! 🧵
www.iambic.ai/post/np3-tec...
if you're interested in the evaluation of conformational accuracy of structure prediction methods, take a look at our first stab at a systematic conformational benchmark in the NP3 technical report below! 🧵
www.iambic.ai/post/np3-tec...

