
Constantin Pape
@cppape.bsky.social
Group leader @ Uni Göttingen for Computational Cell Analytics. Working on AI for biology and medicine with a focus on microscopy image analysis with deep learning.
https://user.informatik.uni-goettingen.de/~pape41/
https://user.informatik.uni-goettingen.de/~pape41/
Hope my counter meme convinces you ;)

November 11, 2025 at 6:13 AM
Hope my counter meme convinces you ;)
The segmentation functionality and other analysis modes, for example for vesicle pool assignments and distance measurements as shown in the screenshot, are available in a napari plugin. Check out github.com/computationa... for details.

October 17, 2025 at 11:09 AM
The segmentation functionality and other analysis modes, for example for vesicle pool assignments and distance measurements as shown in the screenshot, are available in a napari plugin. Check out github.com/computationa... for details.
Are you studying synapses in electron microscopy? Tired of annotating vesicles? We have the tool for you! SynapseNet implements automatic segmentation and analysis of vesicles and other synaptic structures and has now been published:
www.molbiolcell.org/doi/full/10....
www.molbiolcell.org/doi/full/10....

October 17, 2025 at 11:09 AM
Are you studying synapses in electron microscopy? Tired of annotating vesicles? We have the tool for you! SynapseNet implements automatic segmentation and analysis of vesicles and other synaptic structures and has now been published:
www.molbiolcell.org/doi/full/10....
www.molbiolcell.org/doi/full/10....
I did not know Taylor Swift was moonlighting in soliciting contributions for fake journals!
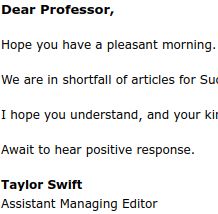
September 16, 2025 at 7:29 PM
I did not know Taylor Swift was moonlighting in soliciting contributions for fake journals!
We released version 1.6 of micro_sam:
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
June 16, 2025 at 3:53 PM
We released version 1.6 of micro_sam:
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
This is how well ChatGPT (o3) knows biology ;)

May 18, 2025 at 6:05 PM
This is how well ChatGPT (o3) knows biology ;)
3. The model selection now supports two additional models (Histopathology, Medical Imaging) and uses human readable names, see the screenshot below.
For more on these models check out: computational-cell-analytics.github.io/micro-sam/mi...
For more on these models check out: computational-cell-analytics.github.io/micro-sam/mi...

March 28, 2025 at 7:08 PM
3. The model selection now supports two additional models (Histopathology, Medical Imaging) and uses human readable names, see the screenshot below.
For more on these models check out: computational-cell-analytics.github.io/micro-sam/mi...
For more on these models check out: computational-cell-analytics.github.io/micro-sam/mi...
2. We added preliminary support for automatic tracking via trackastra, developed by @maweigert.bsky.social.
See the video for an example result and check out computational-cell-analytics.github.io/micro-sam/mi... for details.
See the video for an example result and check out computational-cell-analytics.github.io/micro-sam/mi... for details.
March 28, 2025 at 7:08 PM
2. We added preliminary support for automatic tracking via trackastra, developed by @maweigert.bsky.social.
See the video for an example result and check out computational-cell-analytics.github.io/micro-sam/mi... for details.
See the video for an example result and check out computational-cell-analytics.github.io/micro-sam/mi... for details.
Our next micro_sam release is here! We have a new model for light microscopy, that massively improves for automatic segmentation! See the qualitative and quantitative comparison in the images, v2 is our previous version, v3 is the new one.


February 21, 2025 at 10:36 AM
Our next micro_sam release is here! We have a new model for light microscopy, that massively improves for automatic segmentation! See the qualitative and quantitative comparison in the images, v2 is our previous version, v3 is the new one.
Our first deposition of synaptic vesicles segmentations is now in the Cryo ET Portal! We segmented vesicles in over 50 tomograms to enable analysis of membrane proteins and more.
cryoetdataportal.czscience.com/depositions/...
cryoetdataportal.czscience.com/depositions/...

February 21, 2025 at 6:57 AM
Our first deposition of synaptic vesicles segmentations is now in the Cryo ET Portal! We segmented vesicles in over 50 tomograms to enable analysis of membrane proteins and more.
cryoetdataportal.czscience.com/depositions/...
cryoetdataportal.czscience.com/depositions/...
Our models form the basis of micro_sam, our napari plugin for interactive and automatic segmentation. It can segment data in 2D, 3D and across time.
You can find all the details at github.com/computationa...
You can find all the details at github.com/computationa...
February 12, 2025 at 11:41 AM
Our models form the basis of micro_sam, our napari plugin for interactive and automatic segmentation. It can segment data in 2D, 3D and across time.
You can find all the details at github.com/computationa...
You can find all the details at github.com/computationa...
To achieve these improvements, we finetune SAM on a large LM dataset (ca. 50k images, 1.2M annotated cells) and on a large EM dataset (ca. 5k images, 90k annotated nuclei and mitos). We also add a new decoder for instance segmentation, which provides efficient automatic segmentation.

February 12, 2025 at 11:41 AM
To achieve these improvements, we finetune SAM on a large LM dataset (ca. 50k images, 1.2M annotated cells) and on a large EM dataset (ca. 5k images, 90k annotated nuclei and mitos). We also add a new decoder for instance segmentation, which provides efficient automatic segmentation.
After a long journey, Segment Anything for Microscopy is now published in Nature Methods! We significantly improve SAM for interactive and automatic segmentation in light and electron microscopy and build a user-friendly tool.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
February 12, 2025 at 11:41 AM
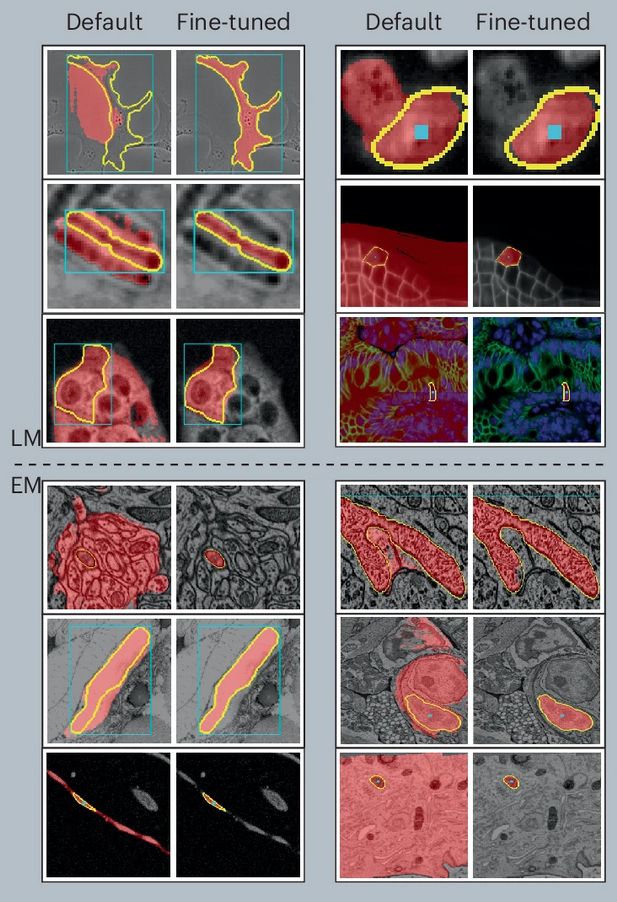
After a long journey, Segment Anything for Microscopy is now published in Nature Methods! We significantly improve SAM for interactive and automatic segmentation in light and electron microscopy and build a user-friendly tool.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
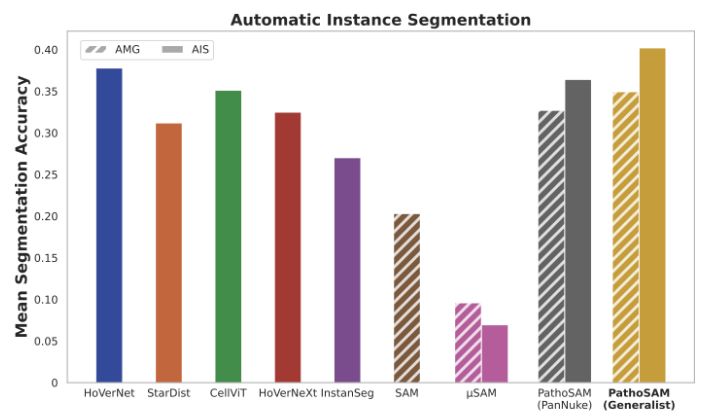
PathoSAM improves SAM forinteractive segmentation of nuclei and adds automatic and semantic segmentation. According to our experiments, it is SOTA for instance segmentation (see ) and performs well for semantic segmentation.
February 11, 2025 at 6:15 AM
PathoSAM improves SAM forinteractive segmentation of nuclei and adds automatic and semantic segmentation. According to our experiments, it is SOTA for instance segmentation (see ) and performs well for semantic segmentation.
Announcing PathoSAM, our foundation model for nucleus segmentation in histopathology. PathoSAM supports interactive and automatic nucleus segmentation (instance and semantic). See segmentation on a WSI from openslide, check out arxiv.org/abs/2502.00408 or read on for details.
February 11, 2025 at 6:15 AM
Announcing PathoSAM, our foundation model for nucleus segmentation in histopathology. PathoSAM supports interactive and automatic nucleus segmentation (instance and semantic). See segmentation on a WSI from openslide, check out arxiv.org/abs/2502.00408 or read on for details.
Our latest preprint PEFT-SAM is on arxiv! We study parameter efficient finetuning to adapt SAM to biomedical images and introduce a new workflow for adaptation based on only two labeled images. See our workflow and improvements in the image, check out arxiv.org/abs/2502.00418 or read on for more.

February 4, 2025 at 11:54 AM
Our latest preprint PEFT-SAM is on arxiv! We study parameter efficient finetuning to adapt SAM to biomedical images and introduce a new workflow for adaptation based on only two labeled images. See our workflow and improvements in the image, check out arxiv.org/abs/2502.00418 or read on for more.
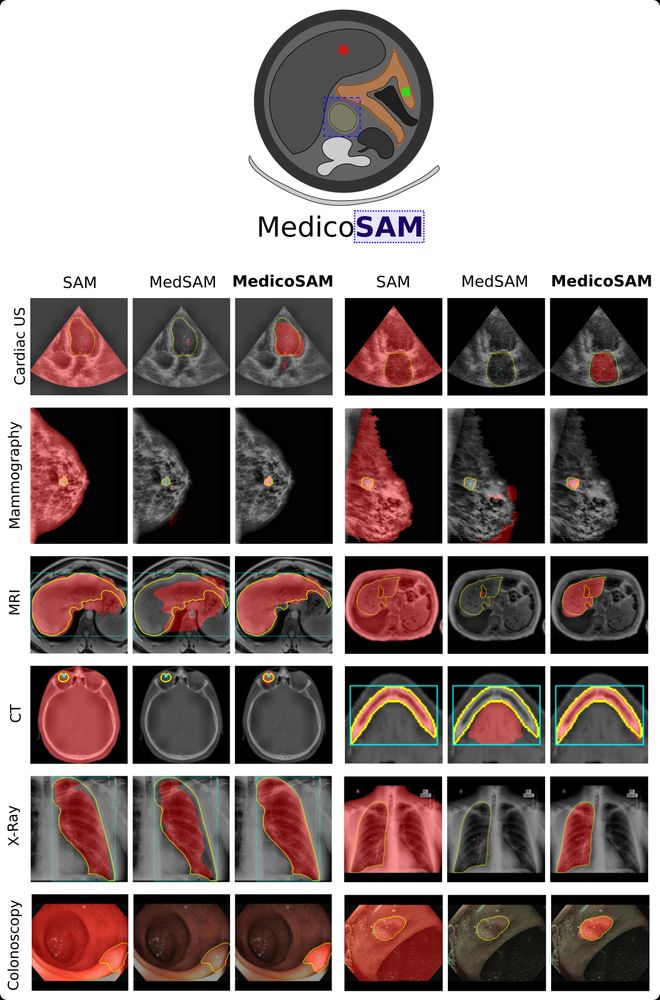
Our work, MedicoSAM, is now on arXiv! We study finetuning SAM for medical images. MedicoSAM improves over SAM and models derived from it, especially for interactive segmentation, see examples in the figure. Preprint at arxiv.org/abs/2501.11734.
Read on for a summary. @anwaiarchit.bsky.social
Read on for a summary. @anwaiarchit.bsky.social
January 22, 2025 at 10:59 AM
Our work, MedicoSAM, is now on arXiv! We study finetuning SAM for medical images. MedicoSAM improves over SAM and models derived from it, especially for interactive segmentation, see examples in the figure. Preprint at arxiv.org/abs/2501.11734.
Read on for a summary. @anwaiarchit.bsky.social
Read on for a summary. @anwaiarchit.bsky.social
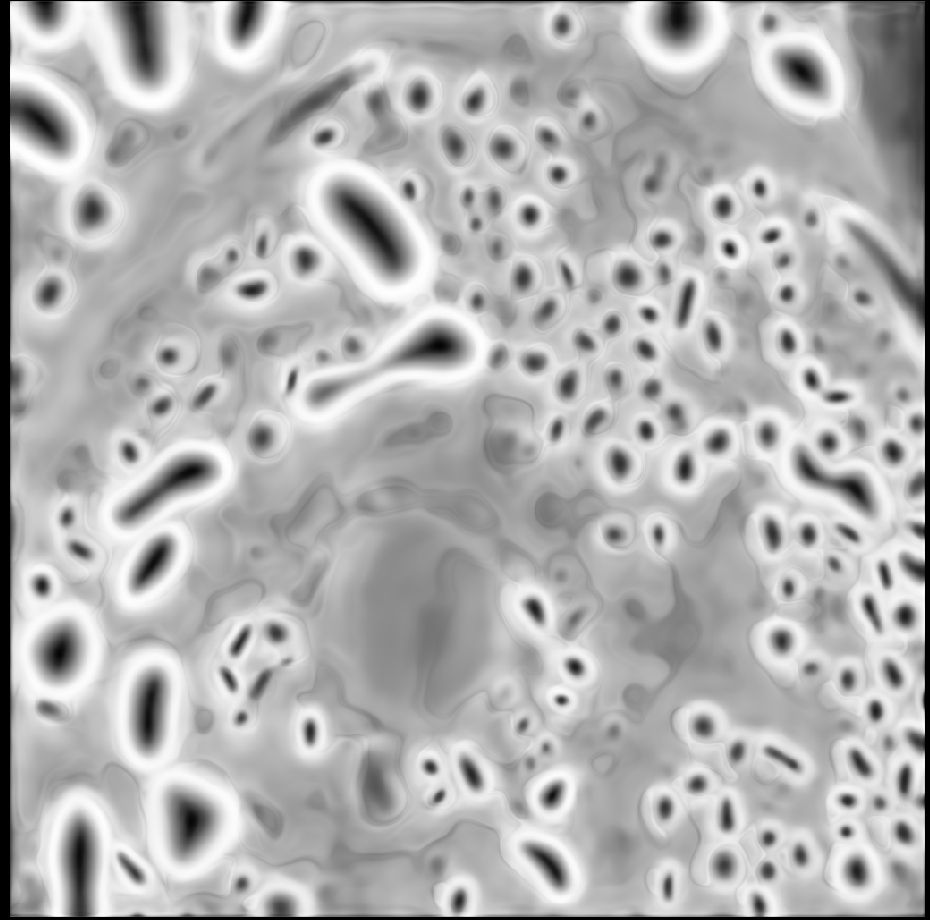
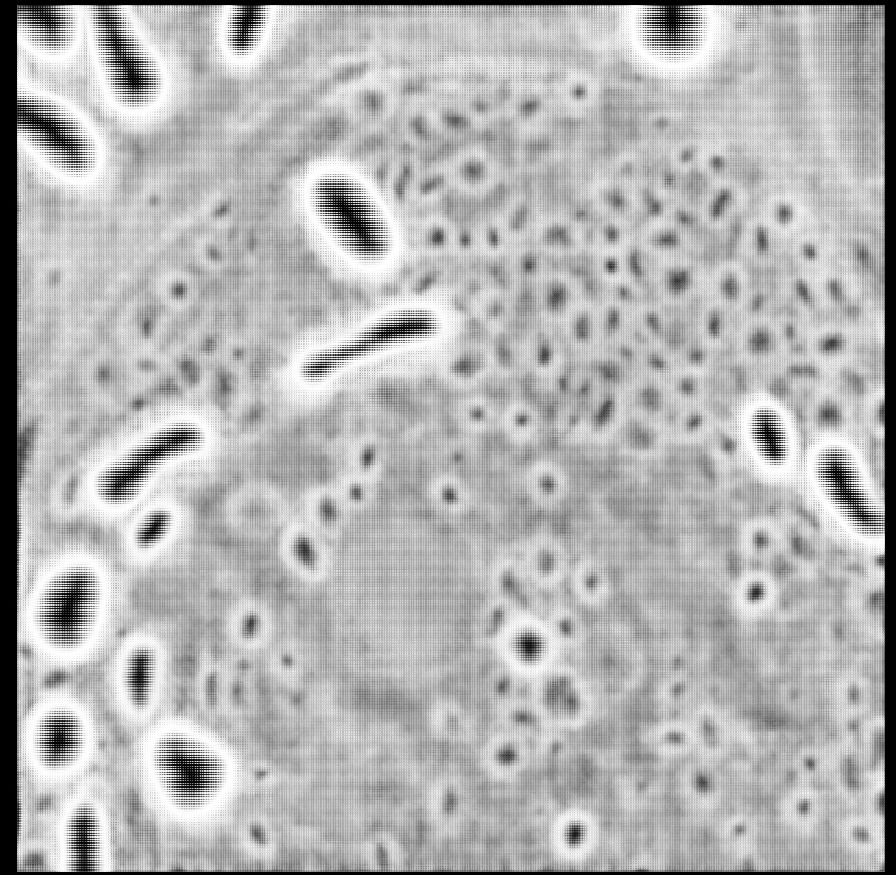
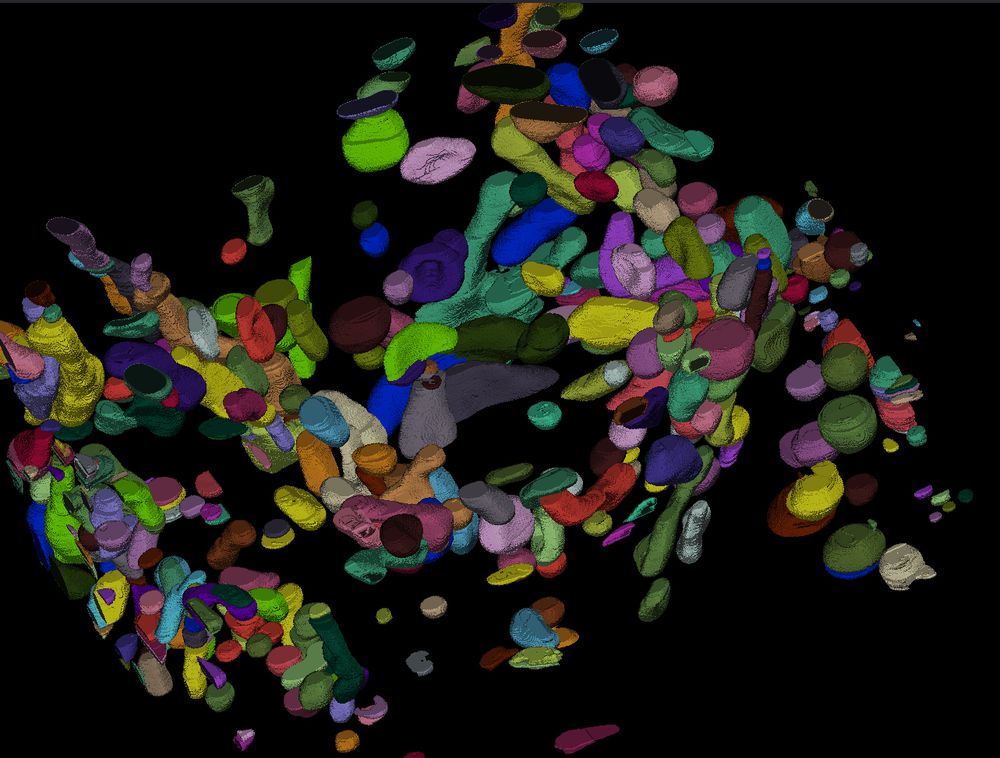
First, to answer the question, according to SynapseNet the electron tomogram of the mossy fibre synapse contains 9,061 vesicles, see the reconstruction of all vesicles rendered in orange. It can also identify active zones (blue), mitochondria (red, cyan) and other synaptic structures.

December 6, 2024 at 3:39 PM
First, to answer the question, according to SynapseNet the electron tomogram of the mossy fibre synapse contains 9,061 vesicles, see the reconstruction of all vesicles rendered in orange. It can also identify active zones (blue), mitochondria (red, cyan) and other synaptic structures.
Want to know how many vesicles there are in a mossy fibre synapse, without counting thousands of vesicles by hand in electron micrographs?
Our new tool SynapseNet has you covered and is now available as a preprint www.biorxiv.org/content/10.1... .
Read on for a short overview of our paper.
Our new tool SynapseNet has you covered and is now available as a preprint www.biorxiv.org/content/10.1... .
Read on for a short overview of our paper.

December 6, 2024 at 3:39 PM
Want to know how many vesicles there are in a mossy fibre synapse, without counting thousands of vesicles by hand in electron micrographs?
Our new tool SynapseNet has you covered and is now available as a preprint www.biorxiv.org/content/10.1... .
Read on for a short overview of our paper.
Our new tool SynapseNet has you covered and is now available as a preprint www.biorxiv.org/content/10.1... .
Read on for a short overview of our paper.

