Label your protein of interest and localize it in cryoET 👁️❄️
Genetically encoded FerriTag as a specific label for cryo-electron tomography: Structure www.cell.com/structure/fu...

Label your protein of interest and localize it in cryoET 👁️❄️
Genetically encoded FerriTag as a specific label for cryo-electron tomography: Structure www.cell.com/structure/fu...
🧬🧊🔬 #Microbiology #CryoEM #PoreFormingToxins
doi.org/10.1101/2025...

🧬🧊🔬 #Microbiology #CryoEM #PoreFormingToxins
doi.org/10.1101/2025...
#ErasmusPlus has received the European Integration Award from the Cercle d’Economia.
🏆 The award honours 35+ years of empowering people, breaking down barriers and uniting Europe through learning and shared values.
To all the changemakers involved, thank you!

#ErasmusPlus has received the European Integration Award from the Cercle d’Economia.
🏆 The award honours 35+ years of empowering people, breaking down barriers and uniting Europe through learning and shared values.
To all the changemakers involved, thank you!
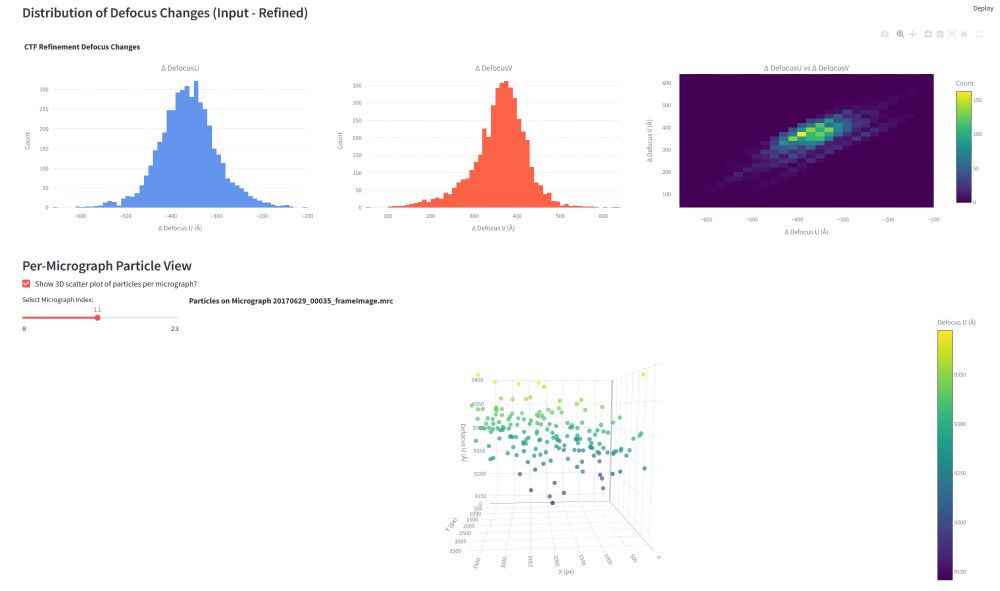
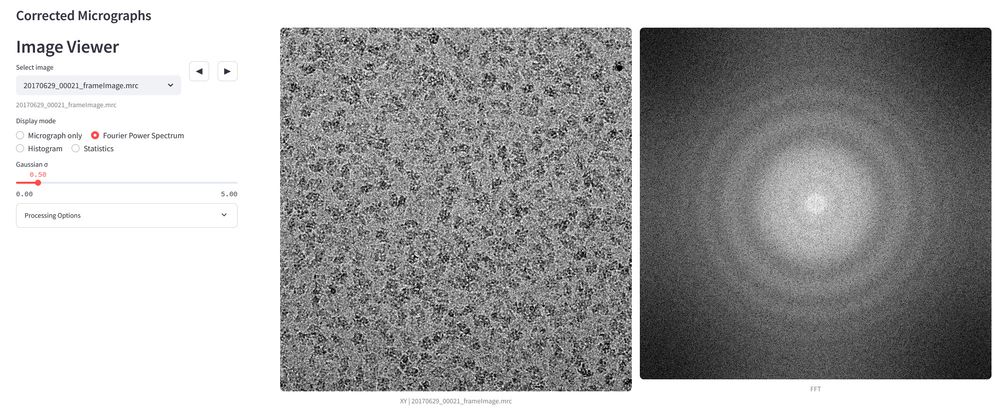
academic.oup.com/bioinformati...



We combine optogenetic stimulation of neurons with in situ cryo-ET to characterize membrane dynamics during synaptic vesicle fusion ❄️🔬⏱️
We combine optogenetic stimulation of neurons with in situ cryo-ET to characterize membrane dynamics during synaptic vesicle fusion ❄️🔬⏱️
@coldpopeye.bsky.social
@unibern.bsky.social
#EPFL #UNIGE #DubochetCenter #MIC
shorturl.at/qyzbb

@coldpopeye.bsky.social
@unibern.bsky.social
#EPFL #UNIGE #DubochetCenter #MIC
shorturl.at/qyzbb
Wonderful coll/w Baumeister lab @madic.bsky.social
#teamtomo #cryoET #cryoEM
Wonderful coll/w Baumeister lab @madic.bsky.social
#teamtomo #cryoET #cryoEM

