github.com/ChrisHIV/tea...

github.com/ChrisHIV/tea...
* the Pandemic Sciences Institute; Data, Epidemiology and Analytics section

* the Pandemic Sciences Institute; Data, Epidemiology and Analytics section

htmlpreview.github.io?https://gith...
I'm very far from an expert in survival analysis - let me know if I said anything silly 😬

htmlpreview.github.io?https://gith...
I'm very far from an expert in survival analysis - let me know if I said anything silly 😬
Read more at 045.medsci.ox.ac.uk/monitoring

Read more at 045.medsci.ox.ac.uk/monitoring







New in Nature: we used digital measurements for 7 million people exposed to confirmed COVID-19 cases to determine risks for virus transmission, captured well by the NHS COVID-19 App. We found that number of hours of exposure is a major predictor, along with proximity.
tinyurl.com/25tfmzee
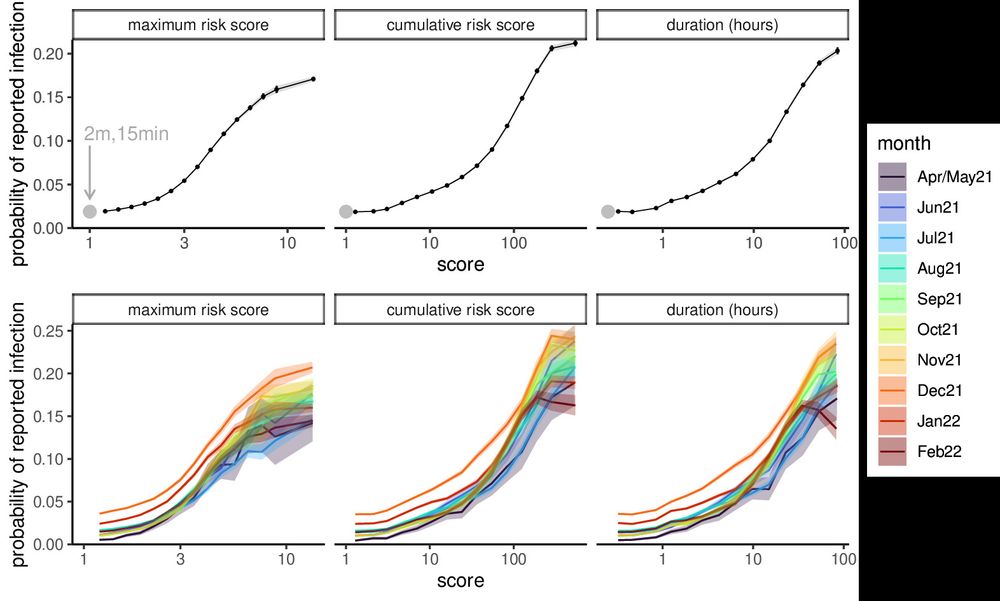
New in Nature: we used digital measurements for 7 million people exposed to confirmed COVID-19 cases to determine risks for virus transmission, captured well by the NHS COVID-19 App. We found that number of hours of exposure is a major predictor, along with proximity.
tinyurl.com/25tfmzee


