www.science.org/doi/10.1126/...
@science.org
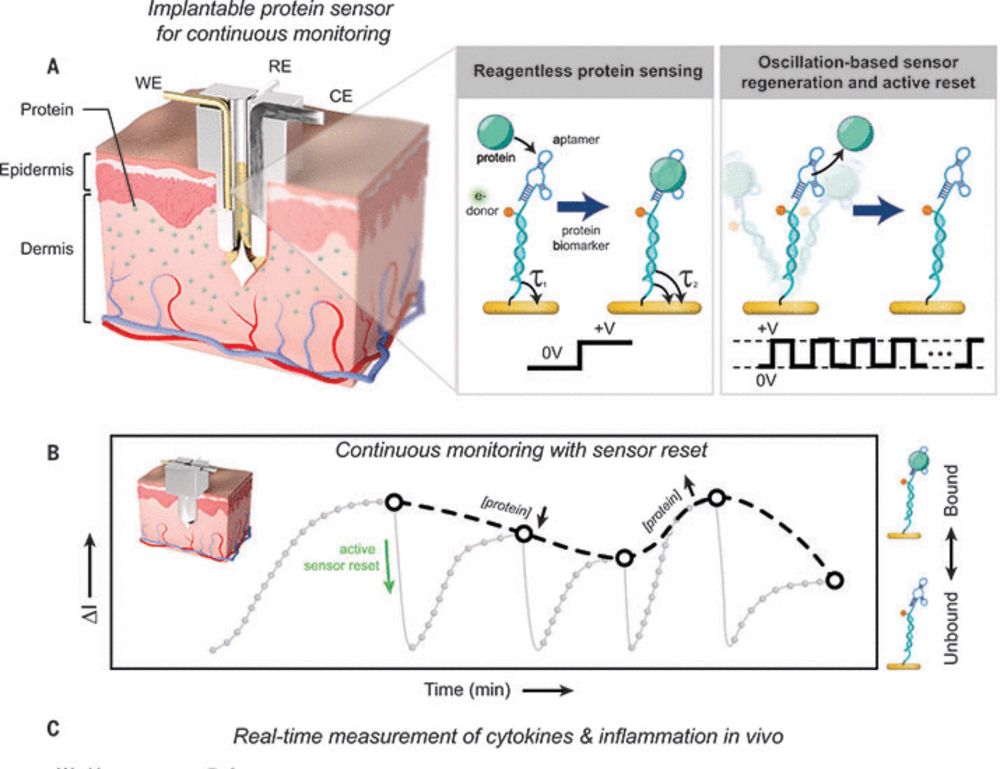
www.science.org/doi/10.1126/...
@science.org
pubs.acs.org/doi/10.1021/...
The key is to understand the biases and factors causing the differences in the data. Those can include and do go beyond the antibodies.
pubs.acs.org/doi/10.1021/...
The key is to understand the biases and factors causing the differences in the data. Those can include and do go beyond the antibodies.
Lack of correlation b/w SomaScan & others might reveal interesting facets of the aptamer tech (protein folding, PTMs, etc) but 100% agree that infighting is bad for the field.
Lack of correlation b/w SomaScan & others might reveal interesting facets of the aptamer tech (protein folding, PTMs, etc) but 100% agree that infighting is bad for the field.

