mysite.science.uottawa.ca/rchica/
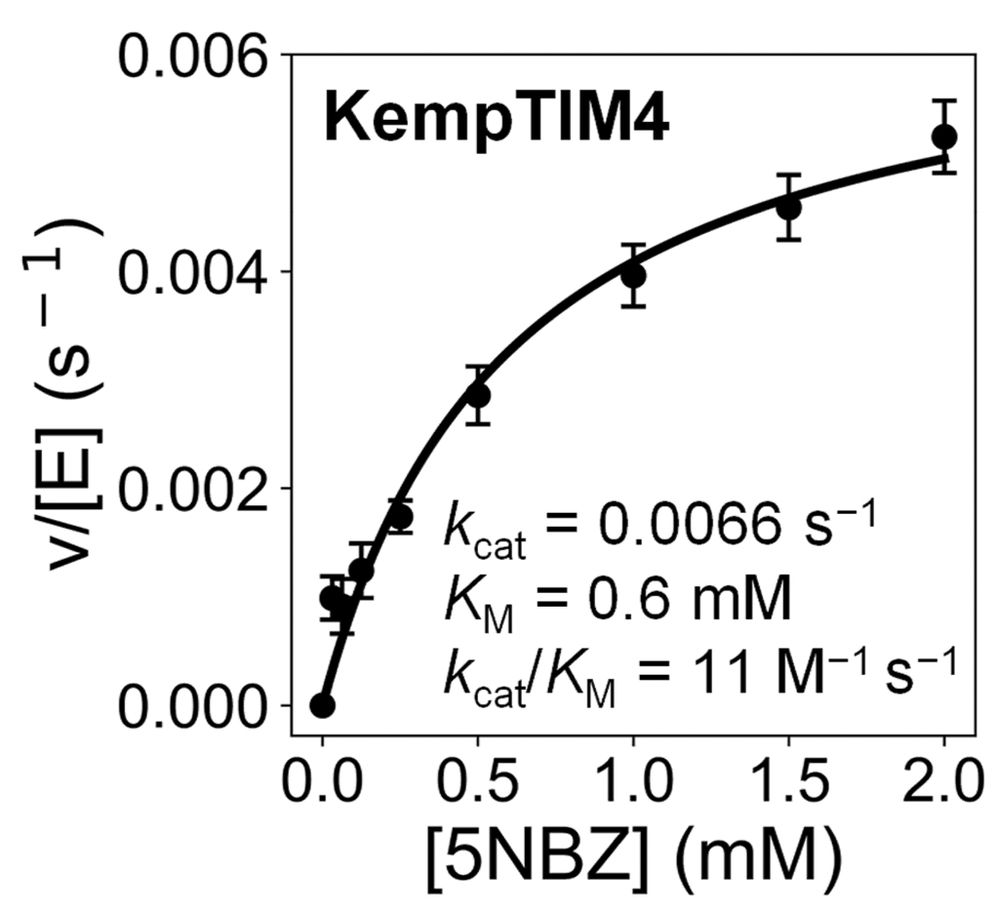
However, a subtle 1.8 Å lid shift disrupted a key catalytic contact, likely contributing to the modest activity. But structural analysis reveals paths to improve activity in the next round of design!
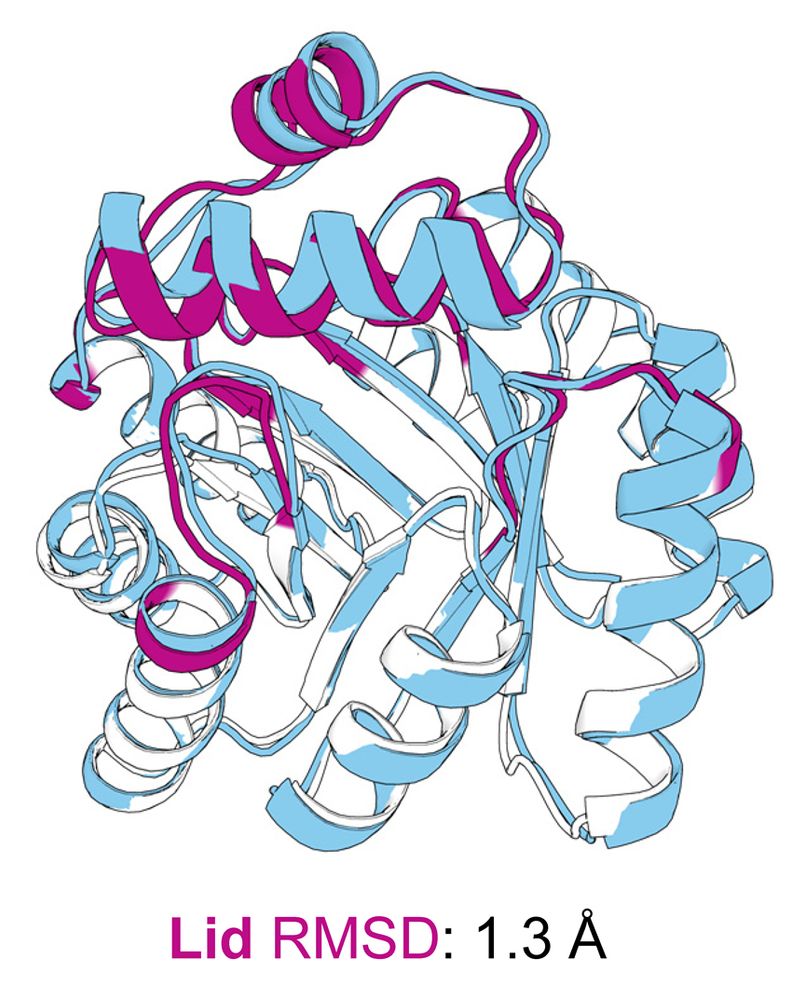
However, a subtle 1.8 Å lid shift disrupted a key catalytic contact, likely contributing to the modest activity. But structural analysis reveals paths to improve activity in the next round of design!
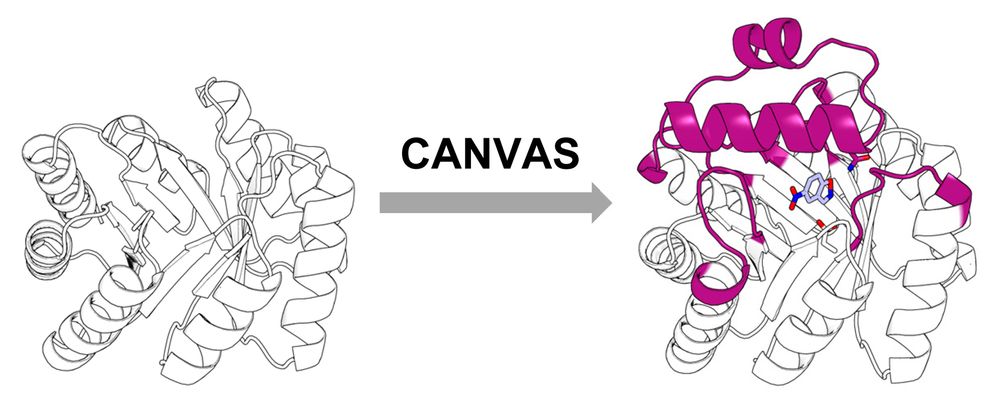
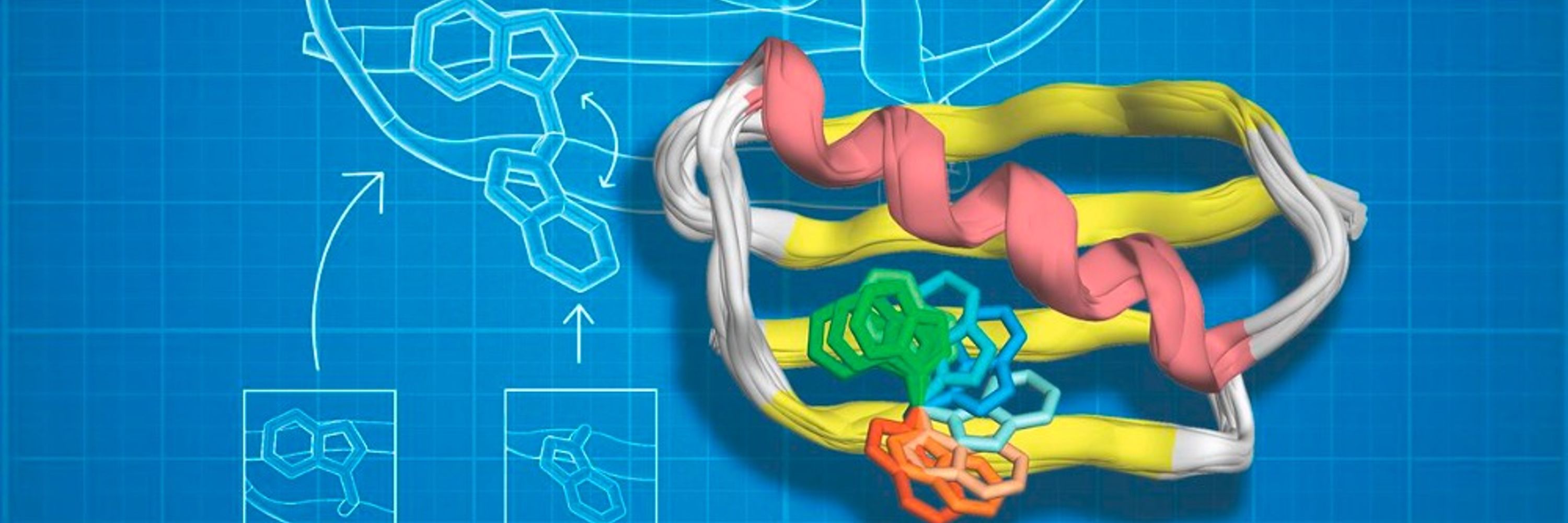
Enter CANVAS: a computational pipeline combining Triad, RFdiffusion & ProteinMPNN to customize minimal TIM barrels into functional enzymes.
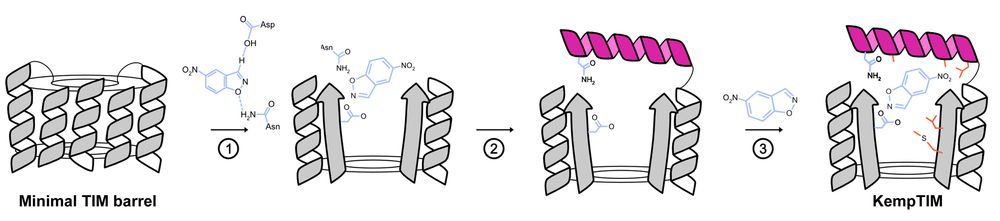
Enter CANVAS: a computational pipeline combining Triad, RFdiffusion & ProteinMPNN to customize minimal TIM barrels into functional enzymes.
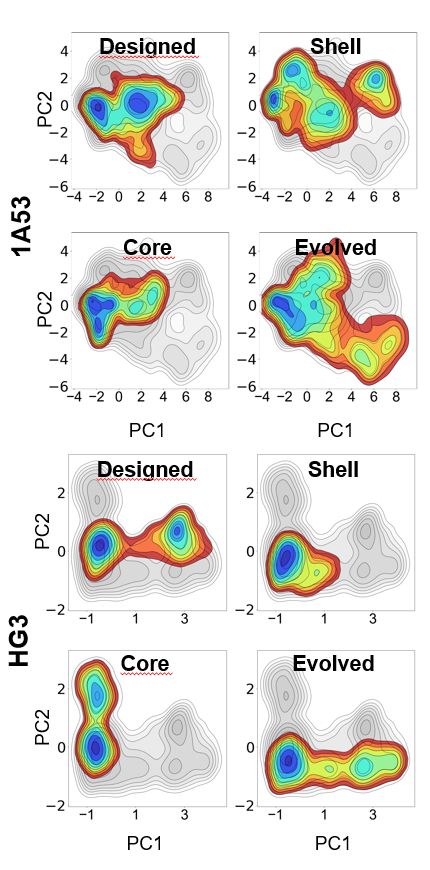

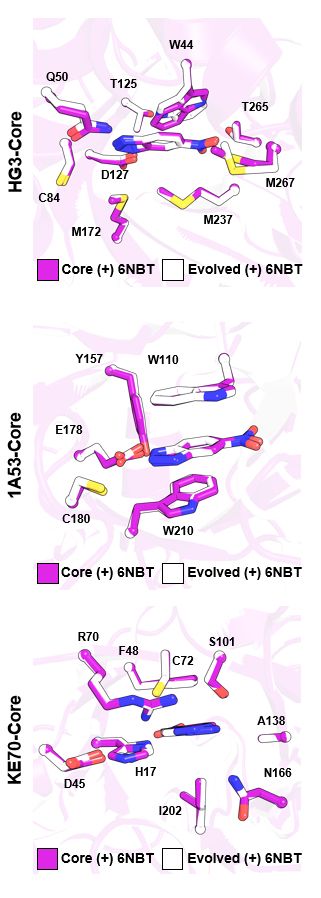
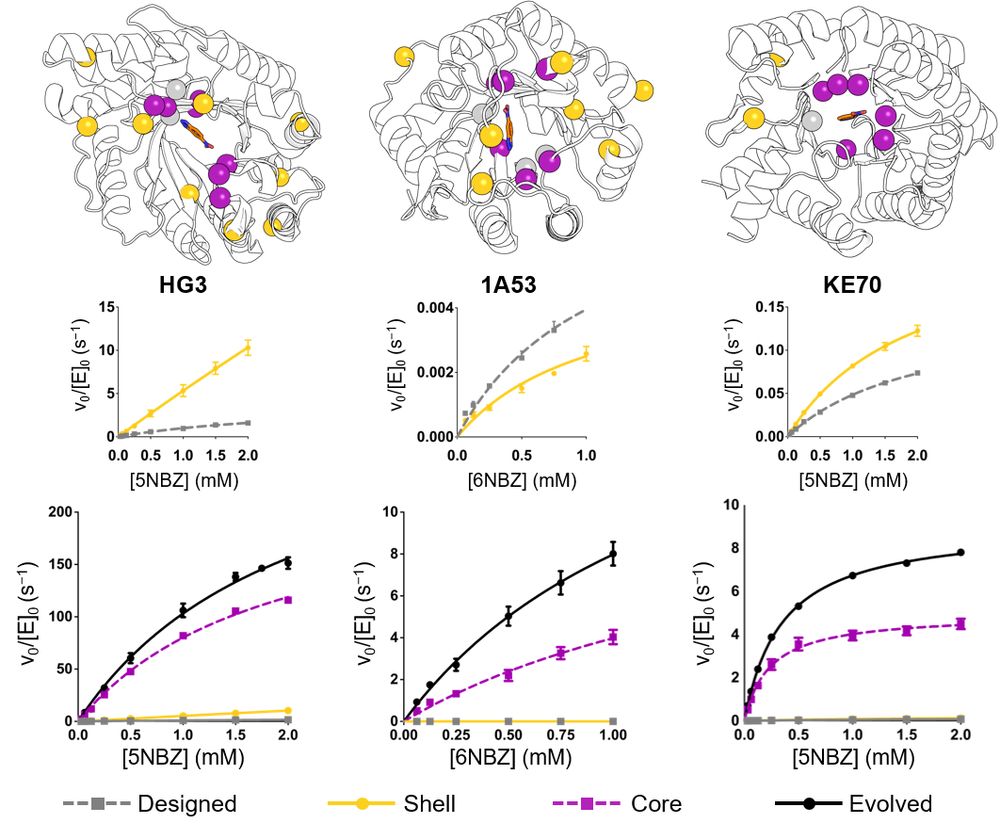
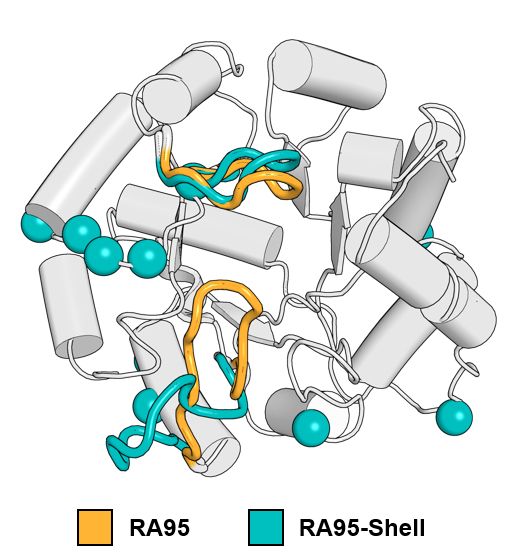
🔹 RA95-Core – Active-site mutations only
🔹 RA95-Shell – Distal mutations only
Comparing them to the evolved RA95.5-8F revealed surprising insights!
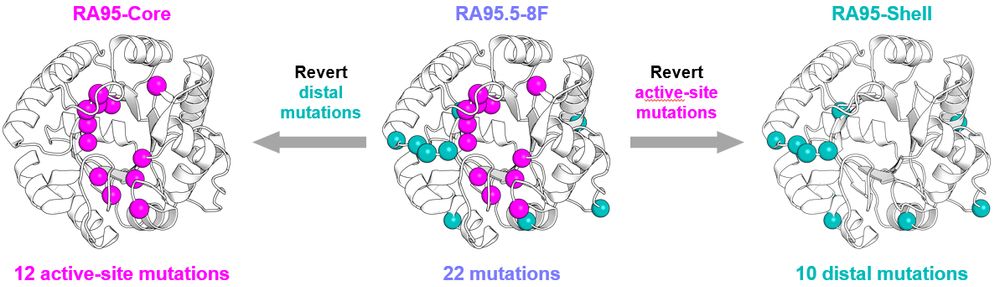
🔹 RA95-Core – Active-site mutations only
🔹 RA95-Shell – Distal mutations only
Comparing them to the evolved RA95.5-8F revealed surprising insights!
1 week left to benefit from early-bird discount on registration!
event.fourwaves.com/pec2024/pages

1 week left to benefit from early-bird discount on registration!
event.fourwaves.com/pec2024/pages

The 5th Protein Engineering Canada Conference will take place at the University of Toronto on June 26-28, 2024.
event.fourwaves.com/pec2024

The 5th Protein Engineering Canada Conference will take place at the University of Toronto on June 26-28, 2024.
event.fourwaves.com/pec2024
The 5th Protein Engineering Canada Conference will take place at the University of Toronto on June 26-28, 2024.
For more info:
event.fourwaves.com/pec2024

The 5th Protein Engineering Canada Conference will take place at the University of Toronto on June 26-28, 2024.
For more info:
event.fourwaves.com/pec2024



