This suggests modern deep learning methods aren’t always better than past methods!

This suggests modern deep learning methods aren’t always better than past methods!
🧪 A standardized evaluation pipeline
🏆 30 challenging motifs as test cases
📊Easy-to-use eval scripts and a leaderboard for method comparison
Now, results can be easily and consistently measured.
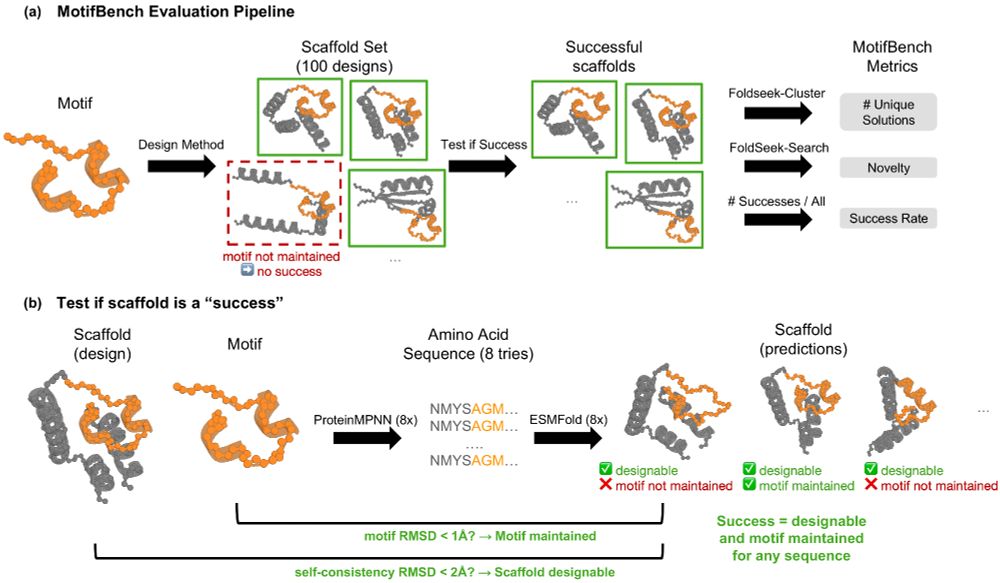
🧪 A standardized evaluation pipeline
🏆 30 challenging motifs as test cases
📊Easy-to-use eval scripts and a leaderboard for method comparison
Now, results can be easily and consistently measured.
✅ Input: a motif (small functional substructure)
🎯 Goal: find a scaffolds (full proteins) that preserves the motif’s geometry.
But what's the state of methods for this problem?
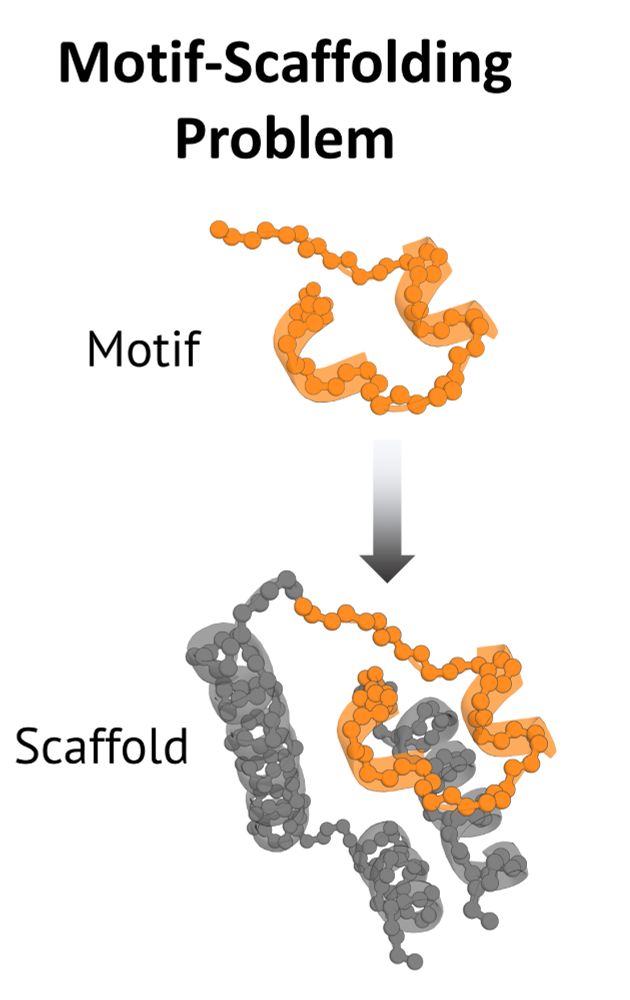
✅ Input: a motif (small functional substructure)
🎯 Goal: find a scaffolds (full proteins) that preserves the motif’s geometry.
But what's the state of methods for this problem?
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
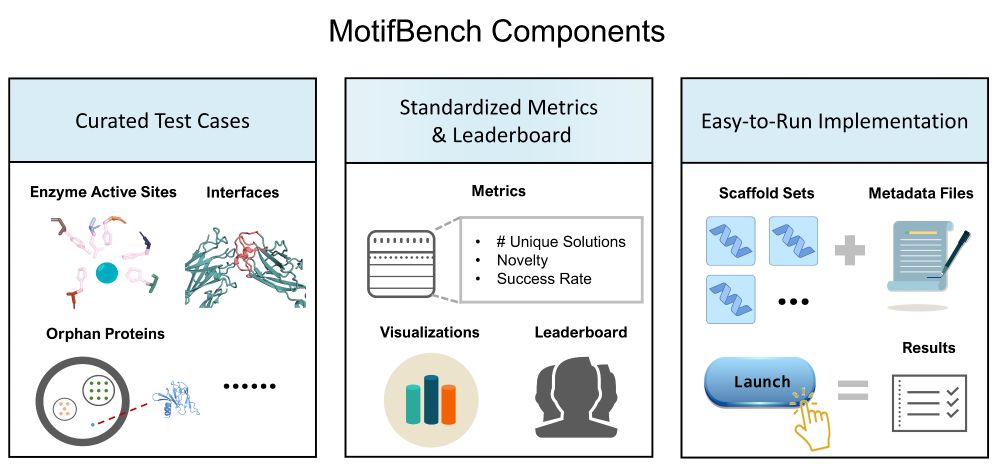
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️

