
Right now I do lots of stats, vis, metabolic modelling and quantitative genomics
can we pls fix greenhouse emissions, gov, pls
39(!) P. aeruginosa strains tested via C. elegans slow-kill assays, followed by GWAS to identify novel accessory genome virulence factors
#MicroSky #Microbiology
doi.org/10.3389/fmic...

39(!) P. aeruginosa strains tested via C. elegans slow-kill assays, followed by GWAS to identify novel accessory genome virulence factors
#MicroSky #Microbiology
doi.org/10.3389/fmic...
In short, using ONLY genotype, one can reasonably predict phenotypic CIP MICs! See Fig 3 (below).
Loving the splitting of MIC and disk diffusion data as well.
#AMR #MicroSky #IDSky

In short, using ONLY genotype, one can reasonably predict phenotypic CIP MICs! See Fig 3 (below).
Loving the splitting of MIC and disk diffusion data as well.
#AMR #MicroSky #IDSky
Impressive benchmarks in Fig 2B (below): ≤10 median errors PER genome
Once again, incredible, landmark work from @rrwick.bsky.social
doi.org/10.1093/bioi...
Tool here: github.com/rrwick/Autoc...
#bioinformatics #genomics

Impressive benchmarks in Fig 2B (below): ≤10 median errors PER genome
Once again, incredible, landmark work from @rrwick.bsky.social
doi.org/10.1093/bioi...
Tool here: github.com/rrwick/Autoc...
#bioinformatics #genomics


#IDSky #MicroSky 🧪
www.biorxiv.org/content/10.1...
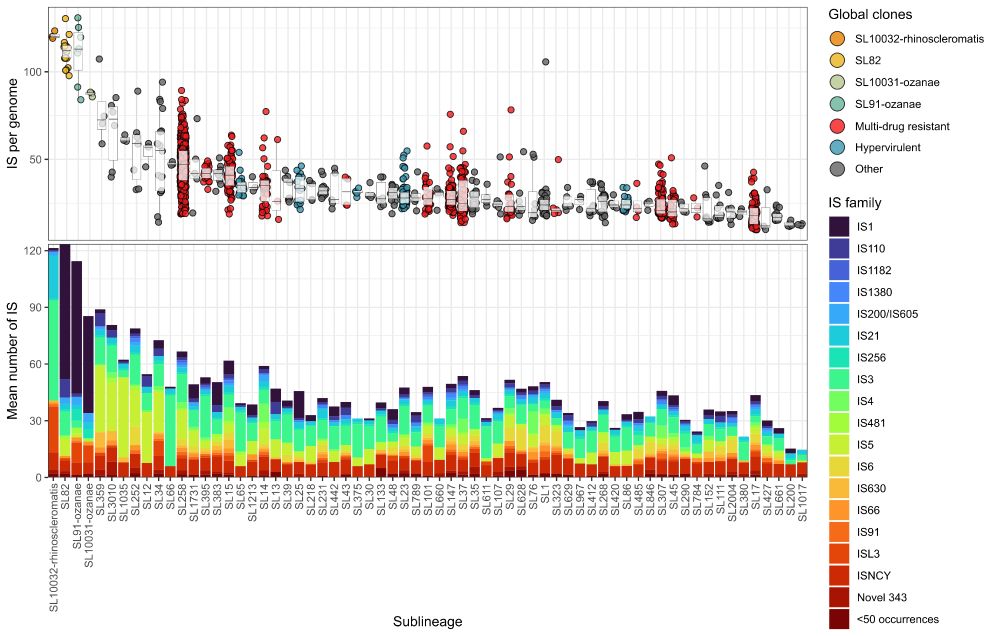
#IDSky #MicroSky 🧪
www.biorxiv.org/content/10.1...
LibreOffice Writer makes formatting easy while Microsoft Word anchors images 5 pages down.
I've made the switch and about to donate. Now LibreOffice Writer is my new best friend ❤️

LibreOffice Writer makes formatting easy while Microsoft Word anchors images 5 pages down.
I've made the switch and about to donate. Now LibreOffice Writer is my new best friend ❤️
The idea is to minimise risk, cost and time at experimental stages while maximising outputs and yield.

The idea is to minimise risk, cost and time at experimental stages while maximising outputs and yield.
www.nobelprize.org/prizes/chemi...

www.nobelprize.org/prizes/chemi...
Long story short: Moon-based PCR. Interesting theory. Strong biochemistry. Weak earth science.
doi.org/10.1016/j.ic...

Long story short: Moon-based PCR. Interesting theory. Strong biochemistry. Weak earth science.
doi.org/10.1016/j.ic...
Capsule blocks conjugation, preventing MPF between donor:recipient. However, conjugation exploits recipient strain heterogeneity of capsule thickness, conjugating to cells with less capsule
#MicroSky 🧪
www.biorxiv.org/content/10.1...
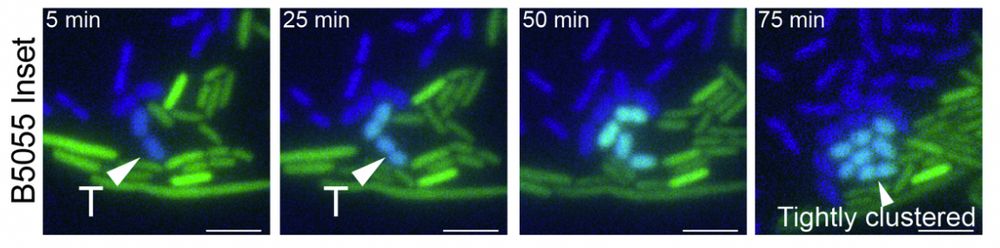
Capsule blocks conjugation, preventing MPF between donor:recipient. However, conjugation exploits recipient strain heterogeneity of capsule thickness, conjugating to cells with less capsule
#MicroSky 🧪
www.biorxiv.org/content/10.1...








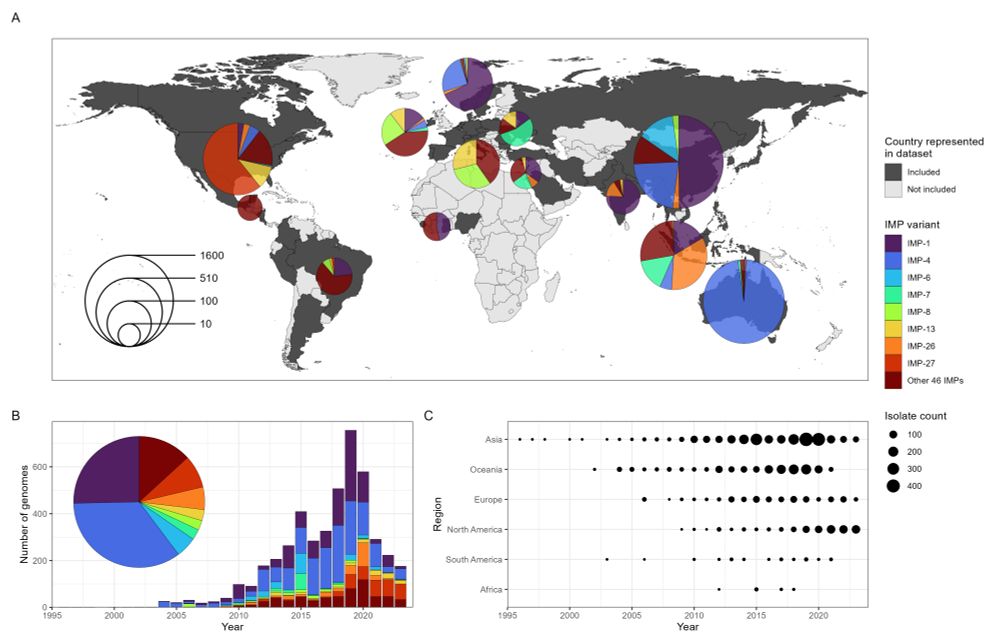
www.medrxiv.org/content/10.1...
#amr #amrsky #IDsky

www.medrxiv.org/content/10.1...
#amr #amrsky #IDsky
Unclear if statistical comparisons controlled for clonal population bias though?
Figure 4C is 👌👌 - using the upper tri for positive associations, and lower tri for negative associations.

Unclear if statistical comparisons controlled for clonal population bias though?
Figure 4C is 👌👌 - using the upper tri for positive associations, and lower tri for negative associations.

