Anna Poetsch
@apoetsch.bsky.social
Computational Biologist at TU Dresden. Interested in how genomes encode their own stability.
Thank you for having me, it was really a very informative, stimulating and motivating few days!
October 1, 2025 at 9:25 AM
Thank you for having me, it was really a very informative, stimulating and motivating few days!
I have already been part of your movement without knowing :-)
September 10, 2025 at 6:38 AM
I have already been part of your movement without knowing :-)
"i think every participant will treasure the experience forever ❤️"
Yes ☺️
Yes ☺️
July 31, 2025 at 6:37 AM
"i think every participant will treasure the experience forever ❤️"
Yes ☺️
Yes ☺️
We have a postdoc position for a project that is very analogous. The deadline has passed, but feel free to still apply, if you are interested. 🧵 6/6

July 31, 2025 at 6:28 AM
We have a postdoc position for a project that is very analogous. The deadline has passed, but feel free to still apply, if you are interested. 🧵 6/6
Such an interdisciplinary interaction also makes ways back to the clinic short, so we hope that these findings can lead to predictive biomarkers for treatment response. Or does modulating NFkB signaling maybe even have potential as a co-treatment strategy 🤔? 🧵 5/6
July 31, 2025 at 6:28 AM
Such an interdisciplinary interaction also makes ways back to the clinic short, so we hope that these findings can lead to predictive biomarkers for treatment response. Or does modulating NFkB signaling maybe even have potential as a co-treatment strategy 🤔? 🧵 5/6
A big shoutout to the Stange Lab at UK Dresden (www.uniklinikum-dresden.de/de/das-klini...), who are building up a very valuable resource with models for gastrointestinal tumours, on which studies like this are based. 🧵 4/6
Stange Lab - Gastrointestinal stem cell and cancer biology
www.uniklinikum-dresden.de
July 31, 2025 at 6:28 AM
A big shoutout to the Stange Lab at UK Dresden (www.uniklinikum-dresden.de/de/das-klini...), who are building up a very valuable resource with models for gastrointestinal tumours, on which studies like this are based. 🧵 4/6
We find changes in the clonal dynamics, but the most striking treatment-related change is the downregulation of inflammatory NFkB signaling in a subpopulation of cells, independent of the genetics of the tumor. This transcriptional memory persists in the organoids without an immune system. 🧵 3/6
July 31, 2025 at 6:28 AM
We find changes in the clonal dynamics, but the most striking treatment-related change is the downregulation of inflammatory NFkB signaling in a subpopulation of cells, independent of the genetics of the tumor. This transcriptional memory persists in the organoids without an immune system. 🧵 3/6
In this paper we used data from patient derived organoids from adenomas of the gastroesophageal junction treated with neoadjuvant FLOT chemotherapy in the patient, or of the organoids from the untreated tumour. We reconstructed the clonal dynamics and integrated the single cell transcriptomes.🧵 2/6

July 31, 2025 at 6:28 AM
In this paper we used data from patient derived organoids from adenomas of the gastroesophageal junction treated with neoadjuvant FLOT chemotherapy in the patient, or of the organoids from the untreated tumour. We reconstructed the clonal dynamics and integrated the single cell transcriptomes.🧵 2/6
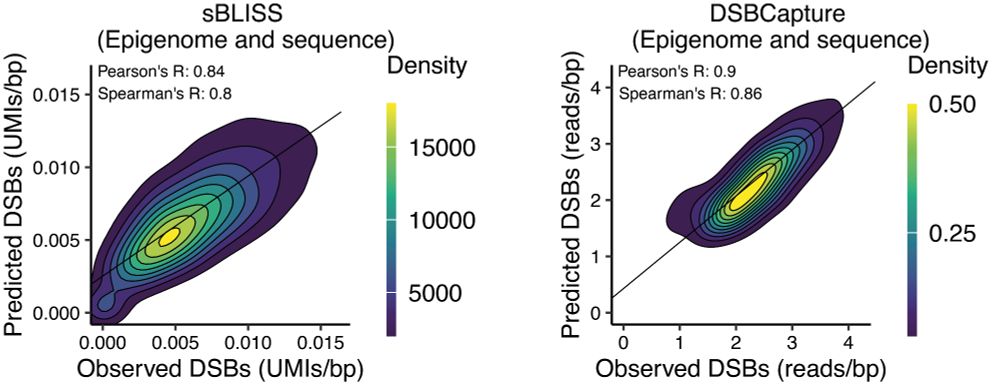
We find that both the 🧬 sequence and the epigenome contribute essential information and the models perform best, when trained on both. But who contributes what? To answer this, interpretable machine learning helps and it is in the paper 😉 🧵 4/4
July 30, 2025 at 2:11 PM
We find that both the 🧬 sequence and the epigenome contribute essential information and the models perform best, when trained on both. But who contributes what? To answer this, interpretable machine learning helps and it is in the paper 😉 🧵 4/4
For this we use our 🧬-language model, GROVER www.nature.com/articles/s42..., and classical machine learning. 🧵 3/4

DNA language model GROVER learns sequence context in the human genome - Nature Machine Intelligence
Genomes can be modelled with language approaches by treating nucleotide bases A, C, G and T like text, but there is no natural concept of what the words would be and whether there is even a ‘language’...
www.nature.com
July 30, 2025 at 2:11 PM
For this we use our 🧬-language model, GROVER www.nature.com/articles/s42..., and classical machine learning. 🧵 3/4

