www.nature.com/articles/s41...
Huge thanks to @cppape.bsky.social for being the absolute best PI, guiding me through every step in this journey, @ritastrack.bsky.social for amazing handling of our paper & all reviewers for helping us strengthen μsam.
Go check our tool and paper right away!😉🧵


www.molbiolcell.org/doi/full/10....

www.molbiolcell.org/doi/full/10....
The paper: arxiv.org/abs/2503.19545 1/🧵

The paper: arxiv.org/abs/2503.19545 1/🧵
www.nature.com/articles/s41...
www.nature.com/articles/s41...
The community is growing and MIDL is becoming the venue-to-go for high quality research discussion!🧵

The community is growing and MIDL is becoming the venue-to-go for high quality research discussion!🧵

The community is growing and MIDL is becoming the venue-to-go for high quality research discussion!🧵
- Improvements for automatic tracking.
- A new experimental mode for object classification.
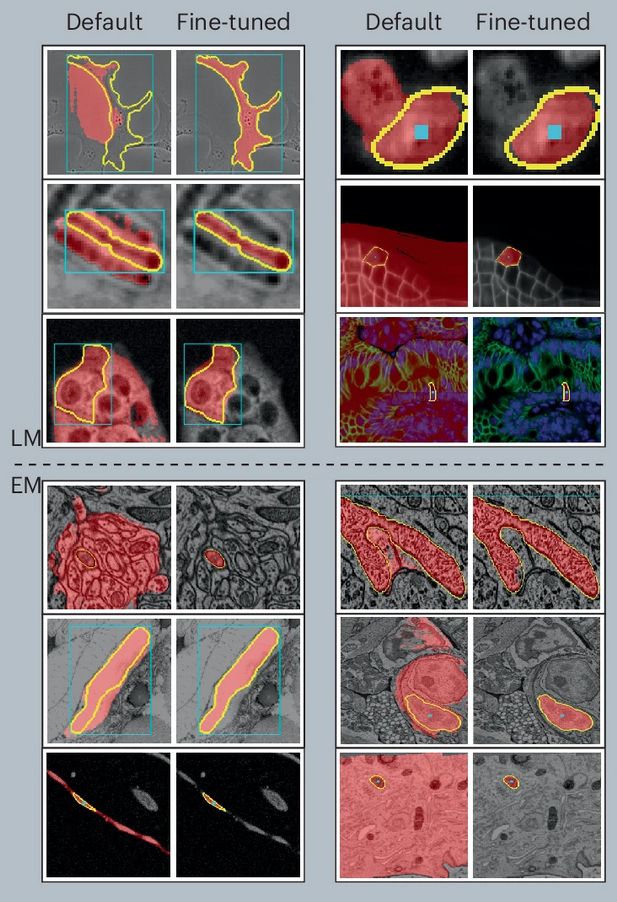
- **New versions of the LM and EM models**
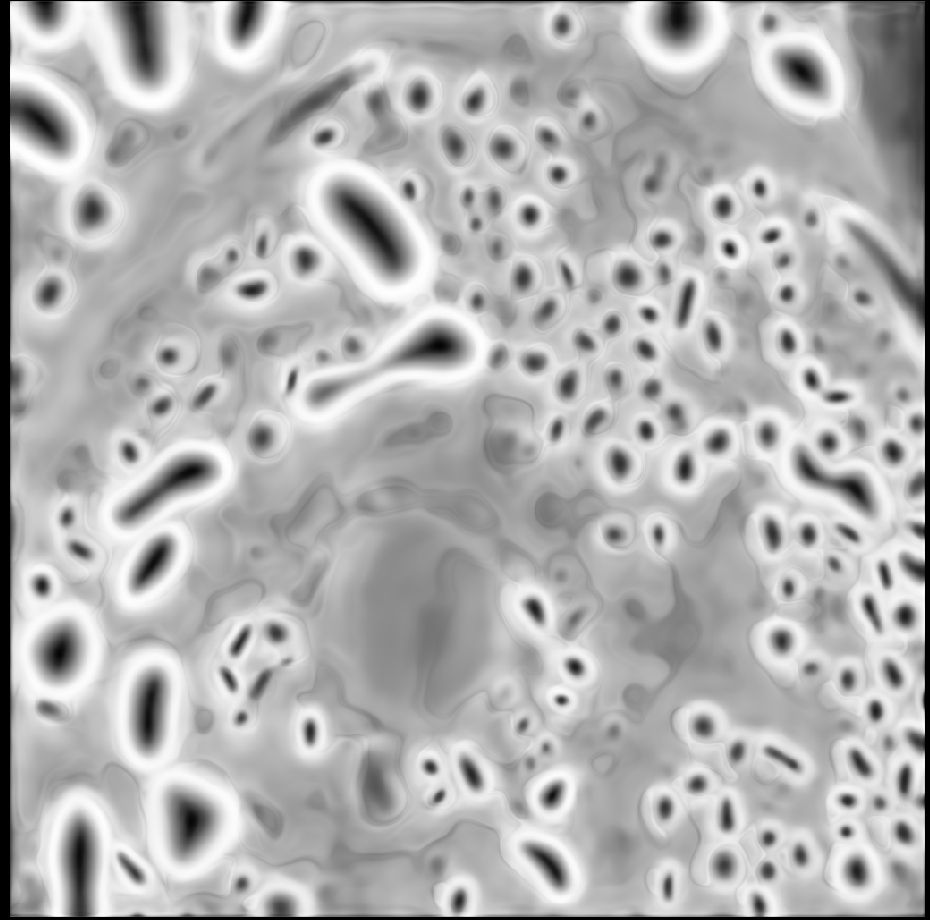
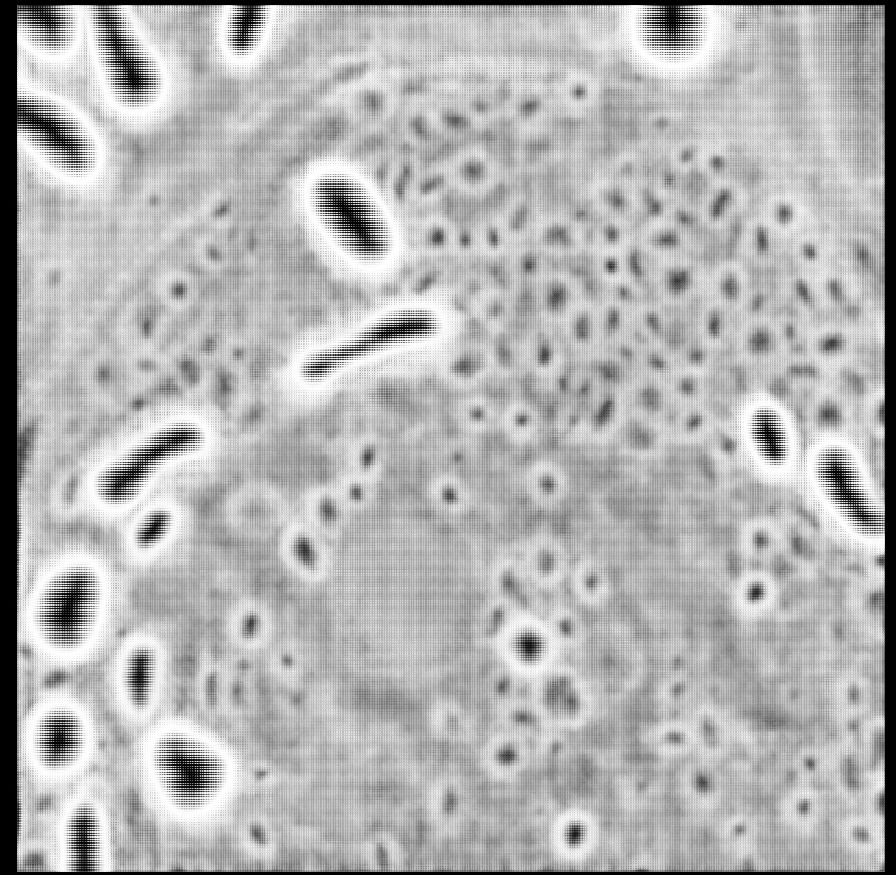
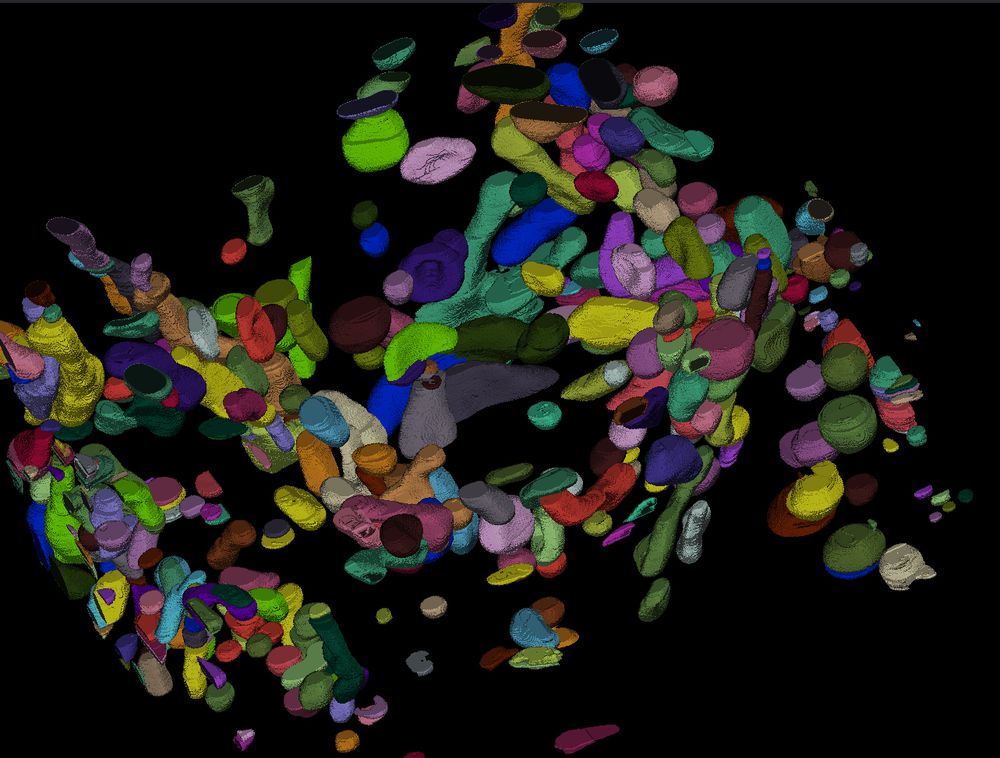
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
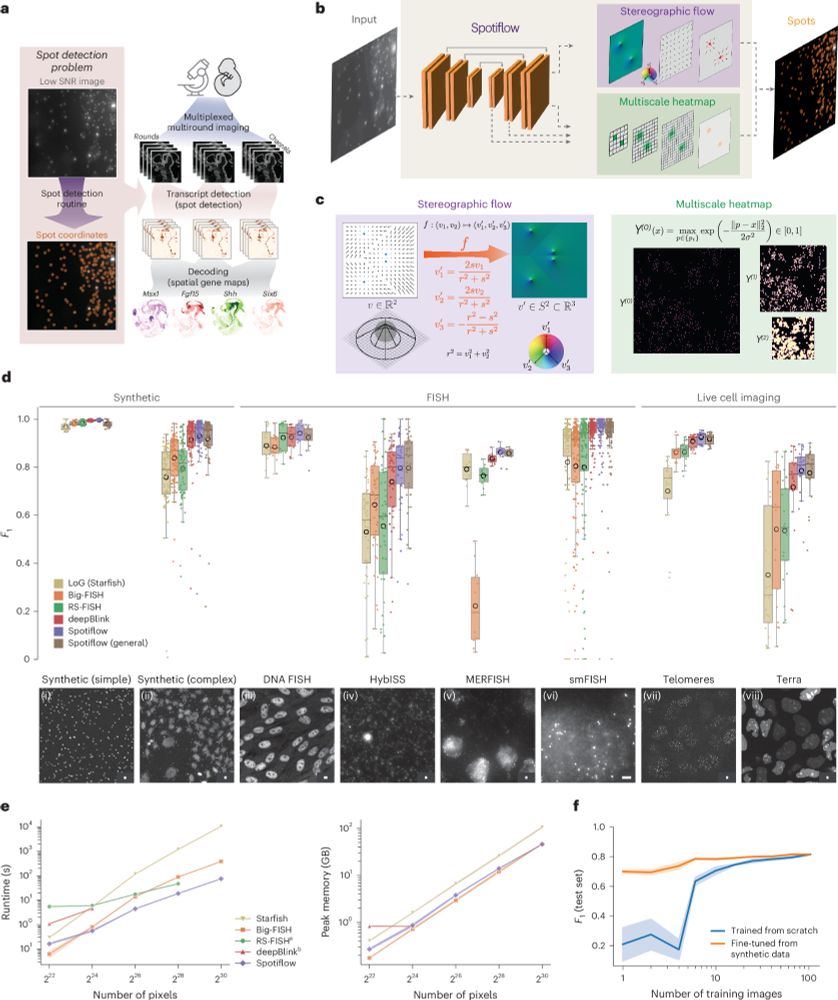
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
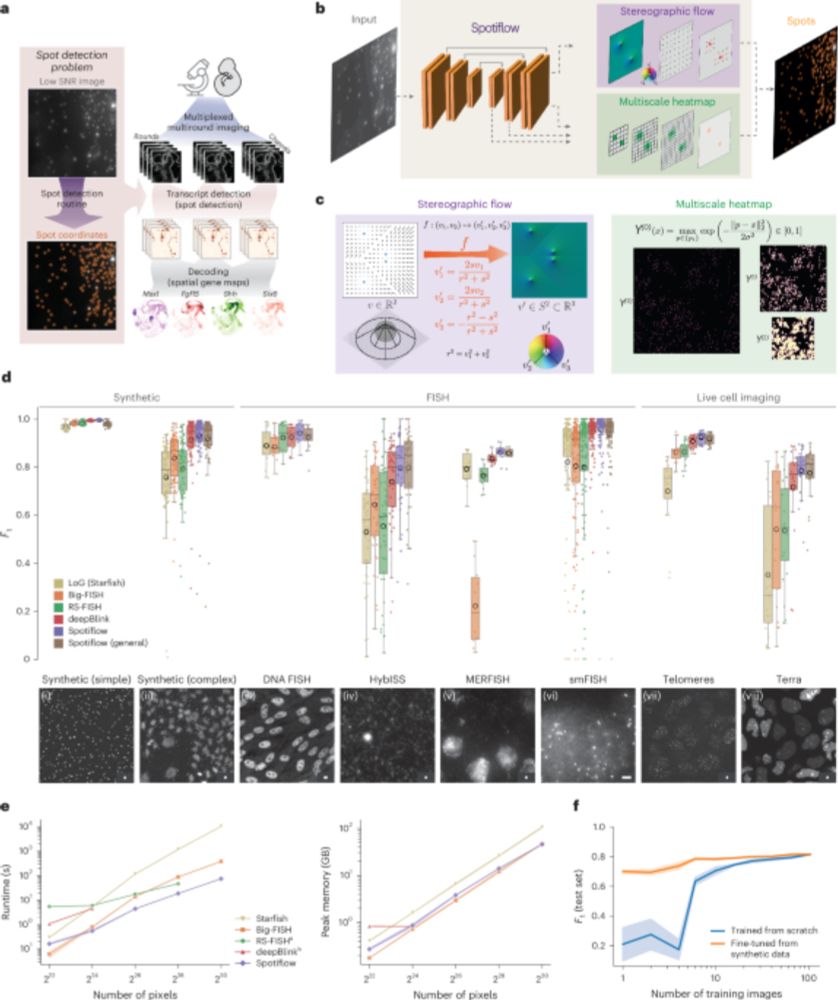
1. Simplified installation on windows.
2. Preliminary support for automatic tracking.
3. Improved interface for model selection.
Read on for a quick summary of the changes.
1. Simplified installation on windows.
2. Preliminary support for automatic tracking.
3. Improved interface for model selection.
Read on for a quick summary of the changes.


nature.com/nmeth/volume...
The cover represents the process of cell and organelle segmentation by Segment Anything for Microscopy.
Paper here: nature.com/articles/s41...
Cover by Sebastian von Haaren.

nature.com/nmeth/volume...
The cover represents the process of cell and organelle segmentation by Segment Anything for Microscopy.
Paper here: nature.com/articles/s41...
Cover by Sebastian von Haaren.
And all thanks to our amazing @haarensv.bsky.social for this! <3
A huge hug to @anwaiarchit.bsky.social and @cppape.bsky.social — it was a pleasure to work alongside you on this!
www.nature.com/nmeth/volume...

And all thanks to our amazing @haarensv.bsky.social for this! <3
Forschungsteam mit bsky.app/profile/cppa...


Forschungsteam mit bsky.app/profile/cppa...
A huge thank you to everyone who attended this advanced course 🙌 We hope the hands-on element of applying deep learning-based methods to your own data and image analysis problems was useful. Until next time!

μsam got some super cool feature updates last week. Don't wait for the next release, go check us out now!
github.com/computationa...
Researchers retrained existing AI-based software on over 17,000 microscopy images with over 2 million structures to develop this new model - Segment Anything for Microscopy: www.uni-goettingen.de/en/3240.html...
#NatureMethods research: doi.org/10.1038/s415...


μsam got some super cool feature updates last week. Don't wait for the next release, go check us out now!
github.com/computationa...


cryoetdataportal.czscience.com/depositions/...

cryoetdataportal.czscience.com/depositions/...
We’re thrilled to announce @ritastrack.bsky.social as a new speaker, delivering an exciting talk on Wednesday. Huge thanks to her for joining us! Stay tuned for more updates on our TiM page: bit.ly/3BPN9fB

We’re thrilled to announce @ritastrack.bsky.social as a new speaker, delivering an exciting talk on Wednesday. Huge thanks to her for joining us! Stay tuned for more updates on our TiM page: bit.ly/3BPN9fB


