doi.org/10.1002/edn3...
Apparently researchers at the University of Zurich conducted an experiment in the /r/changemyview subreddit where they assessed the ability of large language models to change people’s views.
This was allegedly done without any informed consent whatsoever.
Apparently researchers at the University of Zurich conducted an experiment in the /r/changemyview subreddit where they assessed the ability of large language models to change people’s views.
This was allegedly done without any informed consent whatsoever.
besjournals.onlinelibrary.wiley.com/doi/10.1111/...

besjournals.onlinelibrary.wiley.com/doi/10.1111/...
doi.org/10.1002/edn3...

doi.org/10.1002/edn3...
doi.org/10.1002/edn3...

doi.org/10.1002/edn3...
Stay tuned, I'm hoping this one (the manuscript, not the pun) will be an impactful one!
Stay tuned, I'm hoping this one (the manuscript, not the pun) will be an impactful one!

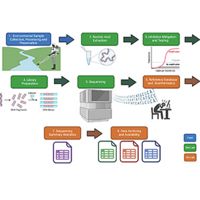
These minimum reporting guidelines for metabarcoding workflows extend from the physical layout of laboratories through to data archiving enhance trust and reliability in data
mbmg.pensoft.net/article/1286...