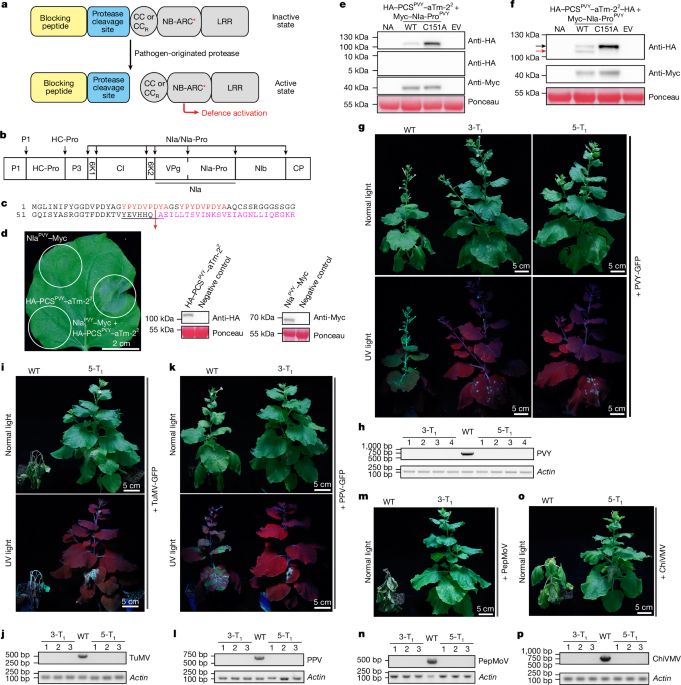
Thank you @sohambio.bsky.social for your persistence! Also to @asimjaved.bsky.social & @jiaxuwu.bsky.social for your contributions! So excited to keep understanding gall-forming pathogens using these results as a starting point!
elifesciences.org/reviewed-pre...
1/n
No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...


"Dasineura asteriae Reprograms the Flower Gene Expressions of Vegetative Organs to Create Flower-Like Gall in Aster scaber"
onlinelibrary.wiley.com/doi/10.1111/...

"Dasineura asteriae Reprograms the Flower Gene Expressions of Vegetative Organs to Create Flower-Like Gall in Aster scaber"
onlinelibrary.wiley.com/doi/10.1111/...
In our new preprint, we describe a new way in which animals and plants share a common strategy to ward off harmful bacteria.
www.biorxiv.org/content/10.1...

In our new preprint, we describe a new way in which animals and plants share a common strategy to ward off harmful bacteria.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...



We cloned AvrWTK4, the first wheat powdery mildew effector recognised by a tandem kinase protein, and show that an HMA-like integrated domain in WTK4 acts as pathogen decoy. Discover the whole story on bioRxiv ⬇️
We cloned AvrWTK4, the first wheat powdery mildew effector recognised by a tandem kinase protein, and show that an HMA-like integrated domain in WTK4 acts as pathogen decoy. Discover the whole story on bioRxiv ⬇️
academic.oup.com/gbe/article/...

academic.oup.com/gbe/article/...
www.nature.com/articles/s41...

The riddle of the enigmatic CCG10-NLR immune receptor family cracked open 🔥
Congrats Guanghao @GuanghaoGuo He Zhang @mhz1989 Selva @M__Selvaraj et al.
#NLRbiology #plantsci #immunology


The riddle of the enigmatic CCG10-NLR immune receptor family cracked open 🔥
Congrats Guanghao @GuanghaoGuo He Zhang @mhz1989 Selva @M__Selvaraj et al.
#NLRbiology #plantsci #immunology
A second paper out today—this time revealing a new source of clubroot resistance in Arabidopsis! 🌱🛡️
Congratulations Melaine González García (@melainegg.bsky.social) 🎉!
Check it out here 👉 apsjournals.apsnet.org/doi/10.1094/...

by Edel Pérez-López (@edelplopez.bsky.social) & colleagues
"If we want meaningful progress...we must critically re-evaluate what qualifies as a truly novel R gene."
FREE access till Oct. 15th here:
authors.elsevier.com/a/1lg2acQbJF...

Thank you @cp-trendsgenetics.bsky.social for a great editorial process!
by Edel Pérez-López (@edelplopez.bsky.social) & colleagues
"If we want meaningful progress...we must critically re-evaluate what qualifies as a truly novel R gene."
FREE access till Oct. 15th here:
authors.elsevier.com/a/1lg2acQbJF...

Thank you @cp-trendsgenetics.bsky.social for a great editorial process!
We identified evolutionary origins of many fungal effectors!
We show that fungi secrete lots of antimicrobial proteins, and that some of them were repurposed by plant pathogens for host immune suppression.
www.biorxiv.org/content/10.1...
cc @teamthomma.bsky.social

We identified evolutionary origins of many fungal effectors!
We show that fungi secrete lots of antimicrobial proteins, and that some of them were repurposed by plant pathogens for host immune suppression.
www.biorxiv.org/content/10.1...
cc @teamthomma.bsky.social
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


📢 #1: Microbiome Diversity and Evolution in Leafhoppers Across Canada
📢 #2: Morphological Changes in Plant Roots Induced by Plasmodiophorid Effectors
📨 To apply submit :
* A cover letter
* Your curriculum vitae
* Academic transcripts
Applications: edelab2024@gmail.com
📢 #1: Microbiome Diversity and Evolution in Leafhoppers Across Canada
📢 #2: Morphological Changes in Plant Roots Induced by Plasmodiophorid Effectors
📨 To apply submit :
* A cover letter
* Your curriculum vitae
* Academic transcripts
Applications: edelab2024@gmail.com
www.nature.com/articles/s41...
Was fun to write this piece with Dina Hochhauser!
Here is a thread to explain the premises
1/
www.nature.com/articles/s41...
Was fun to write this piece with Dina Hochhauser!
Here is a thread to explain the premises
1/
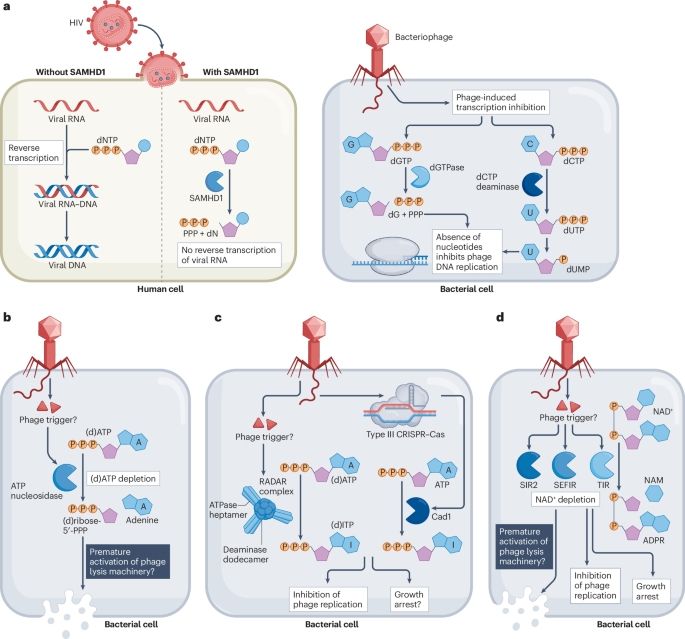
A beautiful example showing how new knowledge on bacterial immunity translates to discoveries important for human health
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
A beautiful example showing how new knowledge on bacterial immunity translates to discoveries important for human health
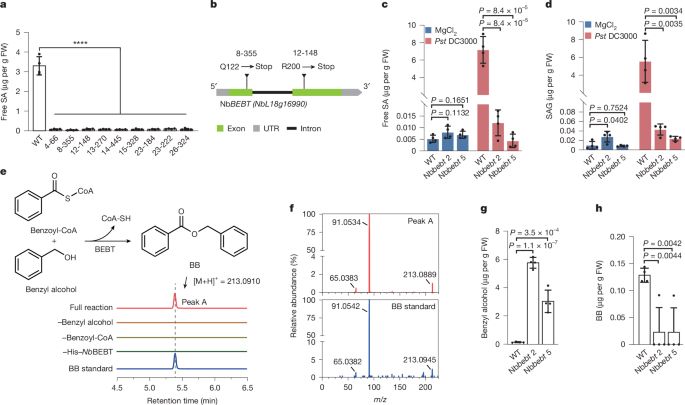
Thank you @cp-trendsmicrobiol.bsky.social for this initiative and for giving visibility to stories that otherwise would have remained hidden 👏
Read @edelplopez.bsky.social and @tatsuyanobori.bsky.social's mobility-related experiences and their insights here: authors.elsevier.com/a/1lTnY,L%7E... & authors.elsevier.com/a/1lTnY_,2Ci...

Thank you @cp-trendsmicrobiol.bsky.social for this initiative and for giving visibility to stories that otherwise would have remained hidden 👏