Ph.D. in Suter’s lab and Fierz’s lab, EPFL
#targetsearch #transcription #singlemoleculeimaging

How do eukaryotic TFs find target sites on the chromatinized genome? How does DNA state—naked or nucleosomal—shape their search strategy?
We investigated how the charge of disordered regions flanking DBDs influences target search and pioneering activity. Check it out! 👇
www.biorxiv.org/content/10.1...
How do eukaryotic TFs find target sites on the chromatinized genome? How does DNA state—naked or nucleosomal—shape their search strategy?
We investigated how the charge of disordered regions flanking DBDs influences target search and pioneering activity. Check it out! 👇
▶️ tinyurl.com/27uxumme
@beatfierz.bsky.social
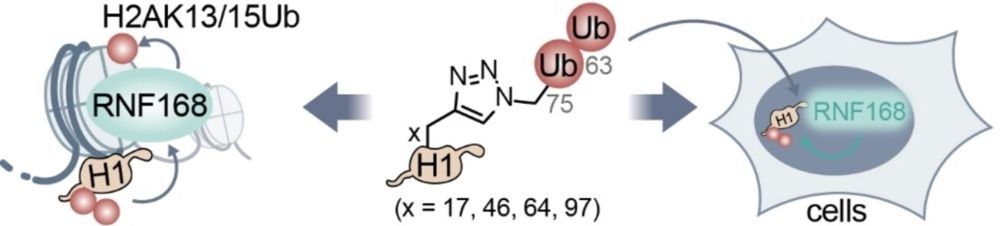
▶️ tinyurl.com/27uxumme
@beatfierz.bsky.social
We just published mechanism-based #cryoEM structures of #SIRT7 on nucleosomes to understand its activity 👇
www.nature.com/articles/s41...
(1/8) #ChemBio #ChemSky
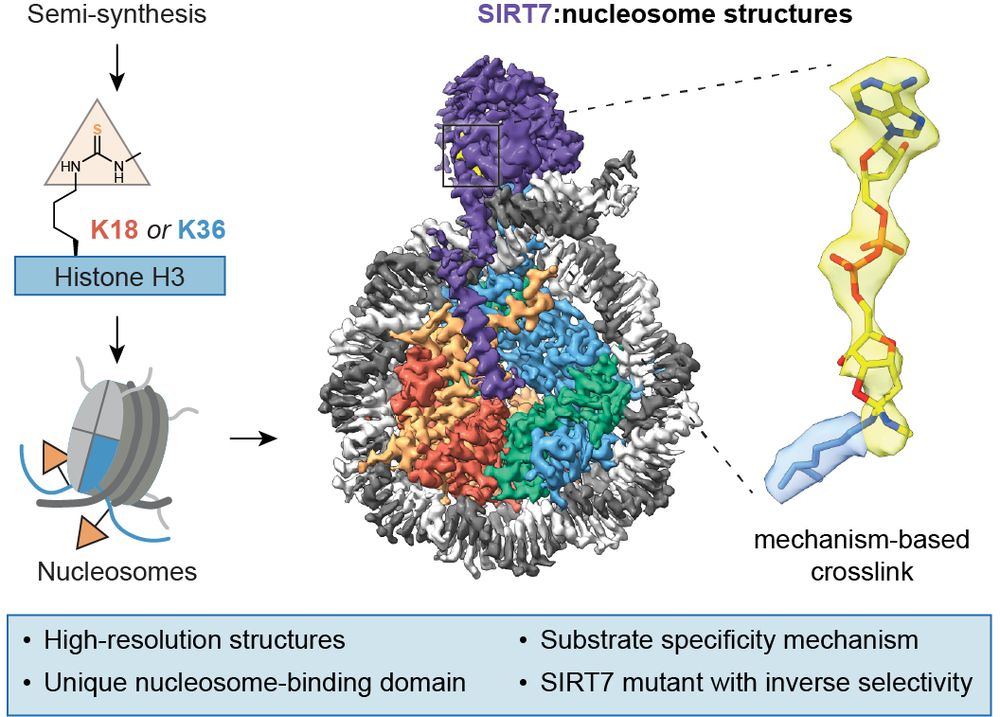
We just published mechanism-based #cryoEM structures of #SIRT7 on nucleosomes to understand its activity 👇
www.nature.com/articles/s41...
(1/8) #ChemBio #ChemSky
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
link.springer.com/protocol/10....

link.springer.com/protocol/10....
Cells walk a tightrope—
proteins made and then destroyed,
balance must be kept.
Yet how is this done?
A mystery in the cell,
we set out to ask.
Live imaging helps,
plus models and omics, too—
data tells the tale.
Cells walk a tightrope—
proteins made and then destroyed,
balance must be kept.
Yet how is this done?
A mystery in the cell,
we set out to ask.
Live imaging helps,
plus models and omics, too—
data tells the tale.
Remodelers and histones,
Who leads, who follows?
🧵 (1/6)
Remodelers and histones,
Who leads, who follows?
🧵 (1/6)
THX to the EMBL travel grant for me to attend such great conference #EMBLOmics

THX to the EMBL travel grant for me to attend such great conference #EMBLOmics

