Ricardo O. Ramirez Flores
@ricoramirez.bsky.social
🇲🇽 Staff Scientist at EMBL-EBI (@ebi.embl.org and @saezlab.bsky.social) studying the molecular biology of multicellular disease processes, cellular cooperation and its parallels to ecology and social systems
Great to see our patient map of heart failure across patient cohorts, scales, and techs out.
Hope that our work shows how we can transform molecular data into interpretable tissue features that describe remodeling and associate with clinical outcomes.
www.nature.com/articles/s41...
Hope that our work shows how we can transform molecular data into interpretable tissue features that describe remodeling and associate with clinical outcomes.
www.nature.com/articles/s41...

A cross-study transcriptional patient map of heart failure defines conserved multicellular coordination in cardiac remodeling - Nature Communications
Cardiac tissue remodeling in heart failure is driven by interactions between multiple cell types, but existing studies have not fully captured these coordinated responses. Here, the authors show that ...
www.nature.com
November 6, 2025 at 1:18 PM
Great to see our patient map of heart failure across patient cohorts, scales, and techs out.
Hope that our work shows how we can transform molecular data into interpretable tissue features that describe remodeling and associate with clinical outcomes.
www.nature.com/articles/s41...
Hope that our work shows how we can transform molecular data into interpretable tissue features that describe remodeling and associate with clinical outcomes.
www.nature.com/articles/s41...
Reposted by Ricardo O. Ramirez Flores
Our revised consensus transcriptional patient map of human heart failure across patient cohorts and single-cell and bulk technologies is now published @natcomms.nature.com www.nature.com/articles/s41...
What are the key disrupted multicellular processes in heart failure? In our new work we combine 23 years of molecular data with recent single-cell atlases to draw a cross-study patient map
doi.org/10.1101/2024...
doi.org/10.1101/2024...

November 6, 2025 at 12:10 PM
Our revised consensus transcriptional patient map of human heart failure across patient cohorts and single-cell and bulk technologies is now published @natcomms.nature.com www.nature.com/articles/s41...
Reposted by Ricardo O. Ramirez Flores
How dit life originate in our planet? How can we create it in the lab?Our @royalsocietypublishing.org Theme Issue "Origins of Life: the possible and the actual", coedited with @sfiscience.bsky.social C Kempes and Susan Stepney is out! royalsocietypublishing.org/toc/rstb/202... @manlius.bsky.social

Philosophical Transactions of the Royal Society B: Biological Sciences: Vol 380, No 1936
royalsocietypublishing.org
October 2, 2025 at 9:04 AM
How dit life originate in our planet? How can we create it in the lab?Our @royalsocietypublishing.org Theme Issue "Origins of Life: the possible and the actual", coedited with @sfiscience.bsky.social C Kempes and Susan Stepney is out! royalsocietypublishing.org/toc/rstb/202... @manlius.bsky.social
Reposted by Ricardo O. Ramirez Flores
Introducing ParTIpy, a python package for Pareto Task Inference that scales to large-scale datasets, including single-cell and spatial transcriptomics.
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

September 15, 2025 at 8:40 AM
Introducing ParTIpy, a python package for Pareto Task Inference that scales to large-scale datasets, including single-cell and spatial transcriptomics.
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
Reposted by Ricardo O. Ramirez Flores
Our Systema framework for evaluating genetic perturbation response prediction methods is now out in @natbiotech.nature.com ✨
Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯
Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯
Systema: a framework for evaluating genetic perturbation response prediction beyond systematic variation - @mariabrbic.bsky.social @mornitzan.bsky.social go.nature.com/4791kda

Systema: a framework for evaluating genetic perturbation response prediction beyond systematic variation - Nature Biotechnology
An evaluation framework isolates perturbation-specific effects in perturbation datasets.
go.nature.com
August 25, 2025 at 6:36 PM
Our Systema framework for evaluating genetic perturbation response prediction methods is now out in @natbiotech.nature.com ✨
Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯
Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯
Reposted by Ricardo O. Ramirez Flores
An analysis shows that current deep learning models do not beat linear baselines in predicting gene perturbation effects, thus emphasizing the importance of further method development and evaluation. @const-ae.bsky.social @wkhuber.bsky.social @s-anders.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
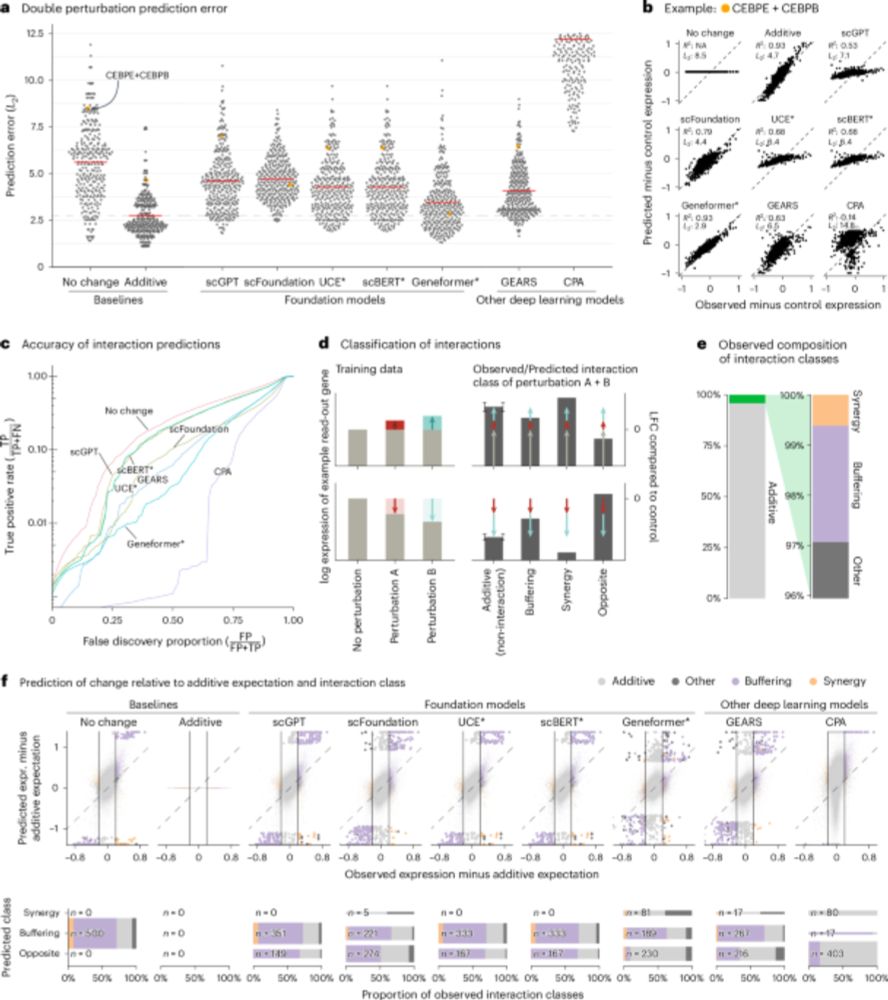
Deep-learning-based gene perturbation effect prediction does not yet outperform simple linear baselines - Nature Methods
The analysis presented in this Brief Communication shows that, despite their complexity, current deep learning models do not outperform linear baselines in predicting gene perturbation effects, thus e...
www.nature.com
August 4, 2025 at 4:07 PM
An analysis shows that current deep learning models do not beat linear baselines in predicting gene perturbation effects, thus emphasizing the importance of further method development and evaluation. @const-ae.bsky.social @wkhuber.bsky.social @s-anders.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Ricardo O. Ramirez Flores
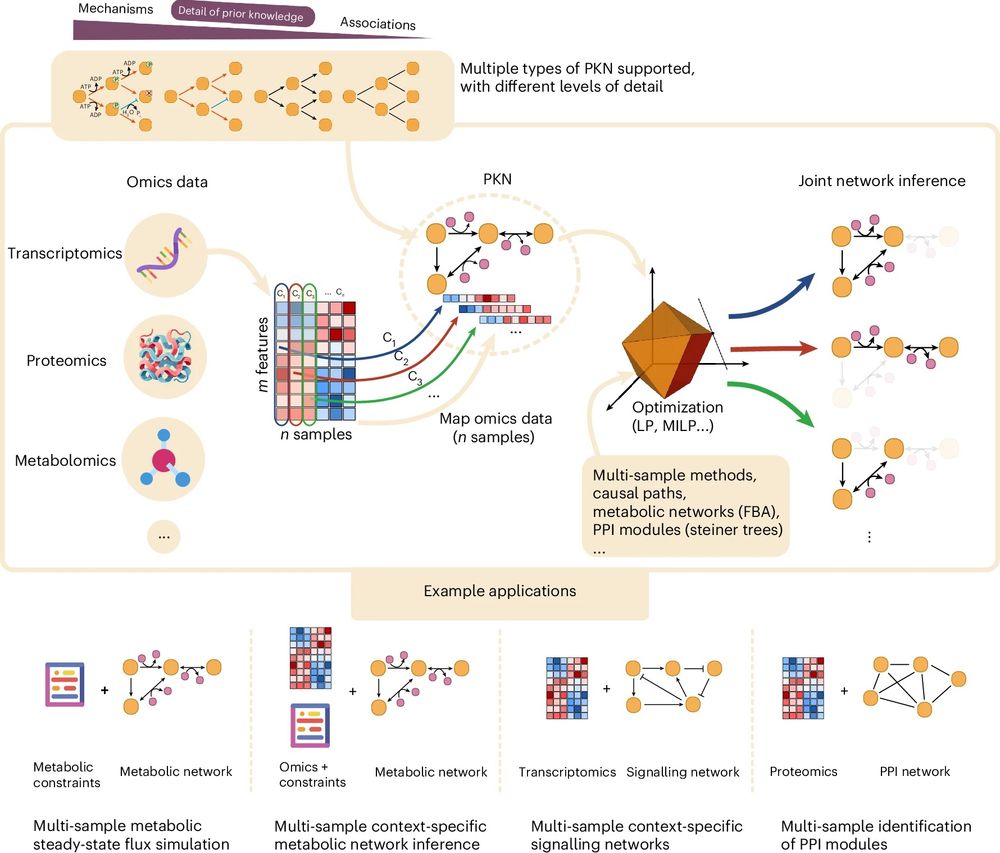
🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is published in peer reviewed form
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
July 22, 2025 at 3:23 PM
🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is published in peer reviewed form
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
Reposted by Ricardo O. Ramirez Flores
From student to researcher, a #career in #science can come with a high price tag. @drcraigmc.bsky.social explores how wealth shapes opportunity in #STEM and proposes structural changes to support #equity and inclusion. 🧪
plos.io/4edGlY4
plos.io/4edGlY4
Too poor to science: How wealth determines who succeeds in STEM
From student to researcher, a career in science can come with a high price tag. This Perspective explores how persistent financial barriers limit who can succeed in science, revealing how wealth shape...
plos.io
June 25, 2025 at 2:39 PM
From student to researcher, a #career in #science can come with a high price tag. @drcraigmc.bsky.social explores how wealth shapes opportunity in #STEM and proposes structural changes to support #equity and inclusion. 🧪
plos.io/4edGlY4
plos.io/4edGlY4
Reposted by Ricardo O. Ramirez Flores
Reposted by Ricardo O. Ramirez Flores
I wanted to write briefly about a very pleasant experience we recently had coordinating and collaborating closely on competing publications with 2 other teams. 1/
January 24, 2025 at 7:36 PM
I wanted to write briefly about a very pleasant experience we recently had coordinating and collaborating closely on competing publications with 2 other teams. 1/
Reposted by Ricardo O. Ramirez Flores
Read preprints. Cite preprints. Email people and tell them you loved their preprint. Email people you hated their preprint. Embrace preprints, preprints are good.
April 22, 2025 at 7:11 PM
Read preprints. Cite preprints. Email people and tell them you loved their preprint. Email people you hated their preprint. Embrace preprints, preprints are good.
Reposted by Ricardo O. Ramirez Flores
The latest version of the Kasumi manuscript is now published in Nature Comms www.nature.com/articles/s41... Kasumi identifies patterns in tissue patches, enabling analysis of disease progression and treatment response while providing insights into spatial coordination at cell-type or marker level
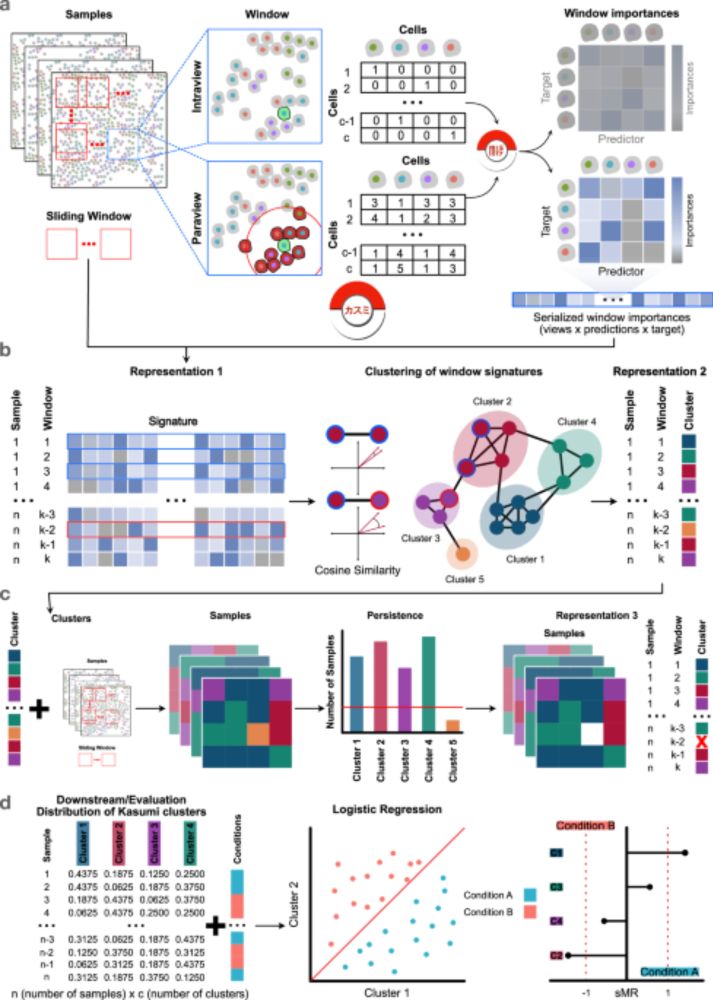
Learning tissue representation by identification of persistent local patterns in spatial omics data - Nature Communications
Spatial omics reveal tissue structures and can aid patient stratification. The authors present a method to identify patterns in tissue patches, enabling analysis of disease progression and treatment r...
www.nature.com
May 7, 2025 at 1:11 PM
The latest version of the Kasumi manuscript is now published in Nature Comms www.nature.com/articles/s41... Kasumi identifies patterns in tissue patches, enabling analysis of disease progression and treatment response while providing insights into spatial coordination at cell-type or marker level
Reposted by Ricardo O. Ramirez Flores
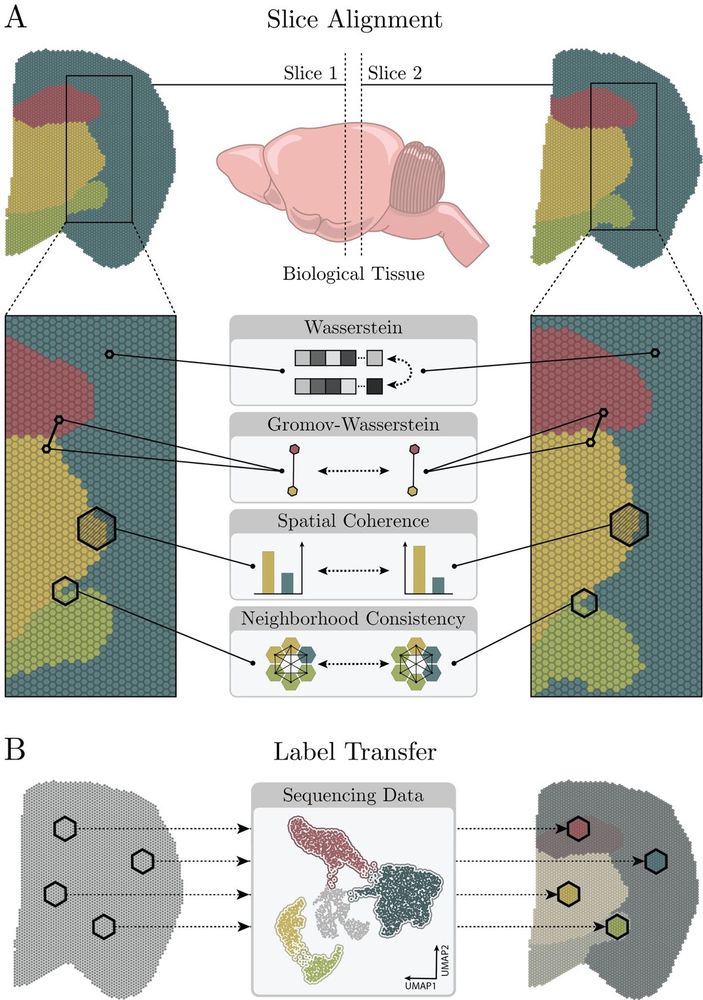
🚨 New preprint: Topography Aware Optimal Transport for Alignment of Spatial Omics Data
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
April 22, 2025 at 9:37 AM
🚨 New preprint: Topography Aware Optimal Transport for Alignment of Spatial Omics Data
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
Reposted by Ricardo O. Ramirez Flores
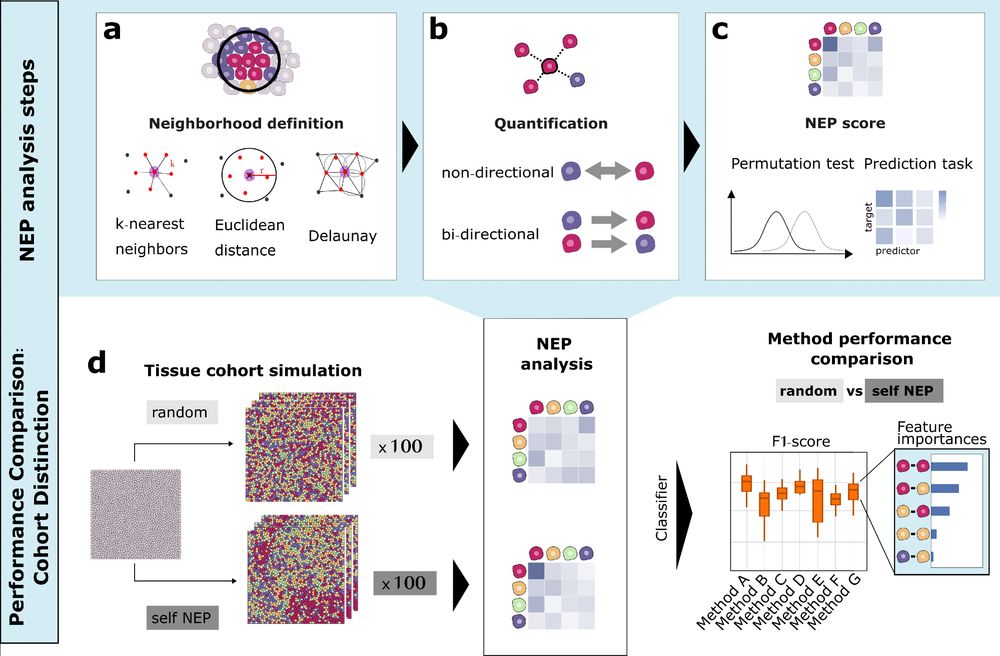
1/
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
April 15, 2025 at 3:50 PM
1/
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
Reposted by Ricardo O. Ramirez Flores
We are excited to be launching the next phase of our Fast & Fair peer review initiative: offering high-quality peer review within 7 working days. #fastandfairpeerreview
Read the Editorial by EiC Daniel Gorelick @danielgorelick.bsky.social at: bit.ly/4kYD1mL
Read the Editorial by EiC Daniel Gorelick @danielgorelick.bsky.social at: bit.ly/4kYD1mL

March 25, 2025 at 11:21 AM
We are excited to be launching the next phase of our Fast & Fair peer review initiative: offering high-quality peer review within 7 working days. #fastandfairpeerreview
Read the Editorial by EiC Daniel Gorelick @danielgorelick.bsky.social at: bit.ly/4kYD1mL
Read the Editorial by EiC Daniel Gorelick @danielgorelick.bsky.social at: bit.ly/4kYD1mL
Reposted by Ricardo O. Ramirez Flores
📄 Update on our preprint about Gene Regulatory Net (GRN) benchmarking 📄
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇

March 14, 2025 at 10:34 AM
📄 Update on our preprint about Gene Regulatory Net (GRN) benchmarking 📄
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇
Reposted by Ricardo O. Ramirez Flores
6 days left to apply to the Post-doc opening in our lab
@ebi.embl.org to develop&apply #bioinformatics & #machine-learning methods to study intra-/extra cellular networks to extract disease mechanisms from #single-cell and #spatial multiomic data: embl.wd103.myworkdayjobs.com/EMBL/job/Hin...
@ebi.embl.org to develop&apply #bioinformatics & #machine-learning methods to study intra-/extra cellular networks to extract disease mechanisms from #single-cell and #spatial multiomic data: embl.wd103.myworkdayjobs.com/EMBL/job/Hin...

Postdoctoral fellow - Saez-Rodriguez Group
Your group Saez-Rodriguez Research Group Your supervisor Julio Saez-Rodriguez Your role As a postdoctoral fellow in the Saez Rodriguez group, you will develop and apply computational methods and tools...
embl.wd103.myworkdayjobs.com
March 17, 2025 at 6:01 PM
6 days left to apply to the Post-doc opening in our lab
@ebi.embl.org to develop&apply #bioinformatics & #machine-learning methods to study intra-/extra cellular networks to extract disease mechanisms from #single-cell and #spatial multiomic data: embl.wd103.myworkdayjobs.com/EMBL/job/Hin...
@ebi.embl.org to develop&apply #bioinformatics & #machine-learning methods to study intra-/extra cellular networks to extract disease mechanisms from #single-cell and #spatial multiomic data: embl.wd103.myworkdayjobs.com/EMBL/job/Hin...
Reposted by Ricardo O. Ramirez Flores
Congrats to @shovalmiyara.bsky.social,Miri Adler, @eldadtzahor.bsky.social & @urialonlab.bsky.social on this great work! We're glad to have supported the translation of findings from theoretical and animal models to human myocardial infarction data using LIANA+ (liana-py.readthedocs.io)
It’s finally here! My PhD work, five years in the making, is now published @CellSystems @cellpress.bsky.social 🚨
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...
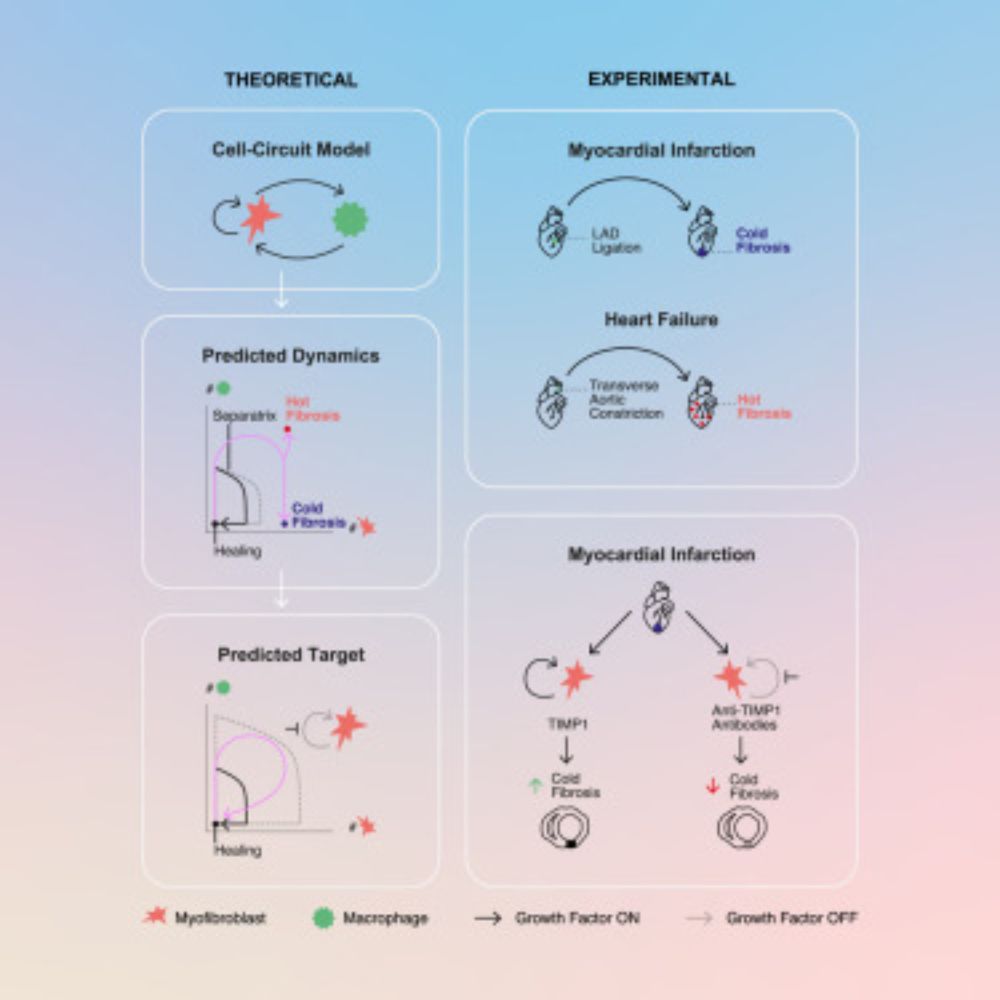
Cold and hot fibrosis define clinically distinct cardiac pathologies
Miyara et al. identify two types of fibrosis in cardiac pathologies: “hot fibrosis,”
involving macrophage-myofibroblast interactions in chronic injuries, and acute-injury-driven
“cold fibrosis,” contr...
www.cell.com
February 25, 2025 at 4:24 PM
Congrats to @shovalmiyara.bsky.social,Miri Adler, @eldadtzahor.bsky.social & @urialonlab.bsky.social on this great work! We're glad to have supported the translation of findings from theoretical and animal models to human myocardial infarction data using LIANA+ (liana-py.readthedocs.io)
Reposted by Ricardo O. Ramirez Flores
It’s finally here! My PhD work, five years in the making, is now published @CellSystems @cellpress.bsky.social 🚨
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...

Cold and hot fibrosis define clinically distinct cardiac pathologies
Miyara et al. identify two types of fibrosis in cardiac pathologies: “hot fibrosis,”
involving macrophage-myofibroblast interactions in chronic injuries, and acute-injury-driven
“cold fibrosis,” contr...
www.cell.com
February 19, 2025 at 3:52 PM
It’s finally here! My PhD work, five years in the making, is now published @CellSystems @cellpress.bsky.social 🚨
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...
Cold and hot fibrosis define clinically distinct cardiac pathologies. www.cell.com/cell-systems...
Reposted by Ricardo O. Ramirez Flores
The first version of NetworkCommons is now published in Bioinformatics. Next, we’ll focus on involving more of the network biology community.
academic.oup.com/bioinformati...
In parallel, we’ll continue expanding benchmarks and developing new applications.
Interested in contributing? Reach out! ⬇️
academic.oup.com/bioinformati...
In parallel, we’ll continue expanding benchmarks and developing new applications.
Interested in contributing? Reach out! ⬇️
February 20, 2025 at 4:30 PM
The first version of NetworkCommons is now published in Bioinformatics. Next, we’ll focus on involving more of the network biology community.
academic.oup.com/bioinformati...
In parallel, we’ll continue expanding benchmarks and developing new applications.
Interested in contributing? Reach out! ⬇️
academic.oup.com/bioinformati...
In parallel, we’ll continue expanding benchmarks and developing new applications.
Interested in contributing? Reach out! ⬇️
Reposted by Ricardo O. Ramirez Flores
JOB OPPORTUNITY: Join us as a scientific programmer to advance tools for multi-omics data at Heidelberg University. Details can be found at shorturl.at/OcnOa and please spread the word!
Scientific programmer
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!
shorturl.at
January 31, 2025 at 8:12 AM
JOB OPPORTUNITY: Join us as a scientific programmer to advance tools for multi-omics data at Heidelberg University. Details can be found at shorturl.at/OcnOa and please spread the word!
Reposted by Ricardo O. Ramirez Flores
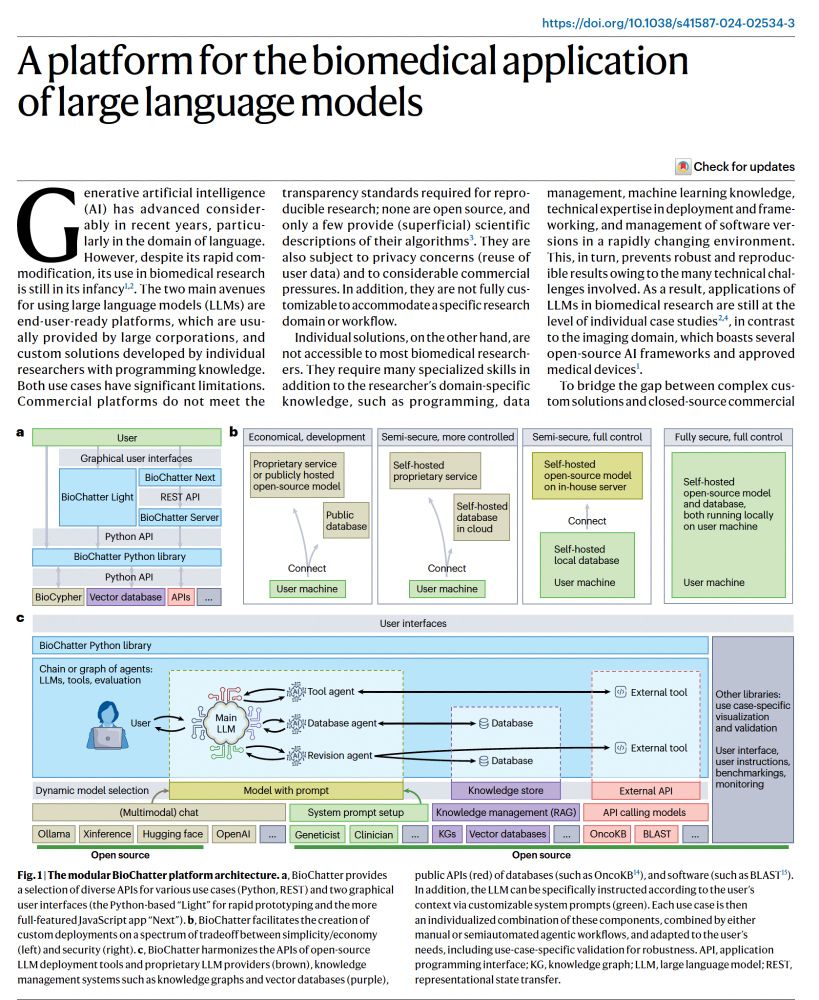
BioChatter, a new open-source platform for large language of life models, "to bridge the gap between complex custom solutions and close-source commercial platforms"
www.nature.com/articles/s41...
@slobentanzer.bsky.social @juliosaezrod.bsky.social
www.nature.com/articles/s41...
@slobentanzer.bsky.social @juliosaezrod.bsky.social
January 22, 2025 at 3:06 PM
BioChatter, a new open-source platform for large language of life models, "to bridge the gap between complex custom solutions and close-source commercial platforms"
www.nature.com/articles/s41...
@slobentanzer.bsky.social @juliosaezrod.bsky.social
www.nature.com/articles/s41...
@slobentanzer.bsky.social @juliosaezrod.bsky.social
Reposted by Ricardo O. Ramirez Flores
1/ 🧵 Free resources for scientists
💡 Covering research skills, careers, #SciComm, leadership, activism and more, everything in this thread appears on our Learning Resources page, with tips for researchers and academics at any career stage. #AcademicChatter #ECRChat
https://buff.ly/4f8vIpk
💡 Covering research skills, careers, #SciComm, leadership, activism and more, everything in this thread appears on our Learning Resources page, with tips for researchers and academics at any career stage. #AcademicChatter #ECRChat
https://buff.ly/4f8vIpk

Learning resources for scientists · eLife
A collections of articles that provide practical resources and guidance for researchers and academics
buff.ly
November 26, 2024 at 3:42 PM
1/ 🧵 Free resources for scientists
💡 Covering research skills, careers, #SciComm, leadership, activism and more, everything in this thread appears on our Learning Resources page, with tips for researchers and academics at any career stage. #AcademicChatter #ECRChat
https://buff.ly/4f8vIpk
💡 Covering research skills, careers, #SciComm, leadership, activism and more, everything in this thread appears on our Learning Resources page, with tips for researchers and academics at any career stage. #AcademicChatter #ECRChat
https://buff.ly/4f8vIpk
Reposted by Ricardo O. Ramirez Flores
Our ChromBPNet preprint out!
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
December 25, 2024 at 11:48 PM
Our ChromBPNet preprint out!
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
Reposted by Ricardo O. Ramirez Flores
Academics from poorer socio-economic backgrounds are more likely to
- not publish
- have outstanding publication records
- introduce more novel scientific concepts
- less likely to receive recognition, as measured by citations, Nobel Prize nominations, and awards.
www.nber.org/papers/w33289
- not publish
- have outstanding publication records
- introduce more novel scientific concepts
- less likely to receive recognition, as measured by citations, Nobel Prize nominations, and awards.
www.nber.org/papers/w33289

December 23, 2024 at 12:10 PM
Academics from poorer socio-economic backgrounds are more likely to
- not publish
- have outstanding publication records
- introduce more novel scientific concepts
- less likely to receive recognition, as measured by citations, Nobel Prize nominations, and awards.
www.nber.org/papers/w33289
- not publish
- have outstanding publication records
- introduce more novel scientific concepts
- less likely to receive recognition, as measured by citations, Nobel Prize nominations, and awards.
www.nber.org/papers/w33289


