QIIME 2
@qiime2.org
Plugin-based biological data science platform that ensures full bioinformatics reproducibility.
Need help? https://forum.qiime2.org
Ready to learn? https://use.qiime2.org
Ready to develop? https://develop.qiime2.org
Need help? https://forum.qiime2.org
Ready to learn? https://use.qiime2.org
Ready to develop? https://develop.qiime2.org
A lot is changing with our documentation, including a transition from a single-source to multiple sources. Learn more in this News post on the forum: forum.qiime2.org/t/the-qiime-...
#qiime2 #bioinformatics #documentation #jupyterbook
#qiime2 #bioinformatics #documentation #jupyterbook

July 2, 2025 at 12:42 PM
A lot is changing with our documentation, including a transition from a single-source to multiple sources. Learn more in this News post on the forum: forum.qiime2.org/t/the-qiime-...
#qiime2 #bioinformatics #documentation #jupyterbook
#qiime2 #bioinformatics #documentation #jupyterbook
Reposted by QIIME 2
I guess I’ll start off on here with my last tweet?
I’m pleased to share my new preprint with @betascience.bsky.social, where we introduce a new database for PICRUSt2 functional predictions.
www.biorxiv.org/content/10.1...
I’m pleased to share my new preprint with @betascience.bsky.social, where we introduce a new database for PICRUSt2 functional predictions.
www.biorxiv.org/content/10.1...

PICRUSt2-MPGA: an update to the reference database used for functional prediction within PICRUSt2
Summary PICRUSt2 is a bioinformatic tool that predicts microbial functions in amplicon sequencing data using a database of annotated reference genomes. We have constructed an updated database for PICR...
www.biorxiv.org
February 20, 2025 at 1:37 PM
I guess I’ll start off on here with my last tweet?
I’m pleased to share my new preprint with @betascience.bsky.social, where we introduce a new database for PICRUSt2 functional predictions.
www.biorxiv.org/content/10.1...
I’m pleased to share my new preprint with @betascience.bsky.social, where we introduce a new database for PICRUSt2 functional predictions.
www.biorxiv.org/content/10.1...
Reposted by QIIME 2
In a spot of happy news, our lab published a new method for kmer-based microbial diversity analysis! The @qiime2.org plugin q2-kmerizer is now out in @asm.org's mSystems 🎉
Check it out #MicroSky and 🖥️🧬 communities interested in #microbiome #bioinformatics:
doi.org/10.1128/msys...
Check it out #MicroSky and 🖥️🧬 communities interested in #microbiome #bioinformatics:
doi.org/10.1128/msys...
Integrating sequence composition information into microbial diversity analyses with k-mer frequency counting | mSystems
k-mers are all of the subsequences of length k that comprise a sequence. Comparing
the frequency of k-mers in DNA sequences yields valuable information about the composition
of these sequences and the...
doi.org
February 23, 2025 at 1:16 PM
In a spot of happy news, our lab published a new method for kmer-based microbial diversity analysis! The @qiime2.org plugin q2-kmerizer is now out in @asm.org's mSystems 🎉
Check it out #MicroSky and 🖥️🧬 communities interested in #microbiome #bioinformatics:
doi.org/10.1128/msys...
Check it out #MicroSky and 🖥️🧬 communities interested in #microbiome #bioinformatics:
doi.org/10.1128/msys...
For the QIIME 2 developers out there, here are the dependency version changes you can expect to see in the upcoming 2025.4 release - reach out if you need help updating your plugins!
forum.qiime2.org/t/qiime-2-20...
forum.qiime2.org/t/qiime-2-20...
QIIME 2 2025.4 breaking changes & environment files
Hello QIIME 2 Developer community! 🤓👋🏼 Below is the list of guaranteed interface changes, plugin API changes, and environment file updates for the 2025.4 QIIME 2 release. You can use the linked envir...
forum.qiime2.org
February 7, 2025 at 5:14 PM
For the QIIME 2 developers out there, here are the dependency version changes you can expect to see in the upcoming 2025.4 release - reach out if you need help updating your plugins!
forum.qiime2.org/t/qiime-2-20...
forum.qiime2.org/t/qiime-2-20...
This announcement is a little late (we're just starting to use bluesky), but #QIIME 2 2024.10 was release this October! Lots of exciting new features with many more still on the way!
forum.qiime2.org/t/qiime-2-20...
#microbiome
#bioinformatics
#microbiomesky
forum.qiime2.org/t/qiime-2-20...
#microbiome
#bioinformatics
#microbiomesky
QIIME 2 2024.10 is now available!
The QIIME 2 2024.10 release is now available! Thanks to everyone involved for their hard work! 🙌🏼 🎉 As a reminder, our next planned QIIME 2 release is scheduled for April 2025 (QIIME 2 2025.4), but p...
forum.qiime2.org
December 12, 2024 at 8:07 PM
This announcement is a little late (we're just starting to use bluesky), but #QIIME 2 2024.10 was release this October! Lots of exciting new features with many more still on the way!
forum.qiime2.org/t/qiime-2-20...
#microbiome
#bioinformatics
#microbiomesky
forum.qiime2.org/t/qiime-2-20...
#microbiome
#bioinformatics
#microbiomesky
Do you use Zenodo for archiving QIIME 2 data for publications? We now support loading QIIME 2 Results (Artifacts and Visualizations) directly from Zenodo with QIIME 2 View.
Check out the links in this Zenodo record, which references data from a new @cap-lab.bio pub:
doi.org/10.5281/zeno...
Check out the links in this Zenodo record, which references data from a new @cap-lab.bio pub:
doi.org/10.5281/zeno...
Upcycling Human Excrement: The Gut Microbiome to Soil Microbiome Axis (supporting data)
This archive contains the supporting data and code for Meilander et al., 2024: Upcycling Human Excrement: The Gut Microbiome to Soil Microbiome Axis. Clicking the links below will open the correspon...
doi.org
November 14, 2024 at 3:33 PM
Do you use Zenodo for archiving QIIME 2 data for publications? We now support loading QIIME 2 Results (Artifacts and Visualizations) directly from Zenodo with QIIME 2 View.
Check out the links in this Zenodo record, which references data from a new @cap-lab.bio pub:
doi.org/10.5281/zeno...
Check out the links in this Zenodo record, which references data from a new @cap-lab.bio pub:
doi.org/10.5281/zeno...
Happy Monday! Start your week off w/ a new QIIME 2 release: @qiime2 2024.2. There's a lot in this one, including new parallel actions in the shotgun distribution, a fancy new visualizer (summarize-plus), for summarizing feature tables & performance enhancements and minor fixes throughout. Enjoy!
QIIME 2 2024.2 is now available!
The QIIME 2 2024.2 release is now available! Thanks to everyone involved for their hard work! 🙌🏼 🎉 As a reminder, our next planned QIIME 2 release is scheduled for May 2023 (QIIME 2 2023.5), but plea...
forum.qiime2.org
February 19, 2024 at 6:16 PM
Happy Monday! Start your week off w/ a new QIIME 2 release: @qiime2 2024.2. There's a lot in this one, including new parallel actions in the shotgun distribution, a fancy new visualizer (summarize-plus), for summarizing feature tables & performance enhancements and minor fixes throughout. Enjoy!
Reposted by QIIME 2
QIIME 2 Provenance Replay auto-generates the code for creating QIIME 2 results from QIIME 2 results & builds "bioinformatics reproducibility supplements" for your papers. Our goal is to make it easy for you to perform reproducible, replicable & robust bioinformatics.
doi.org/10.1371/jour...
doi.org/10.1371/jour...
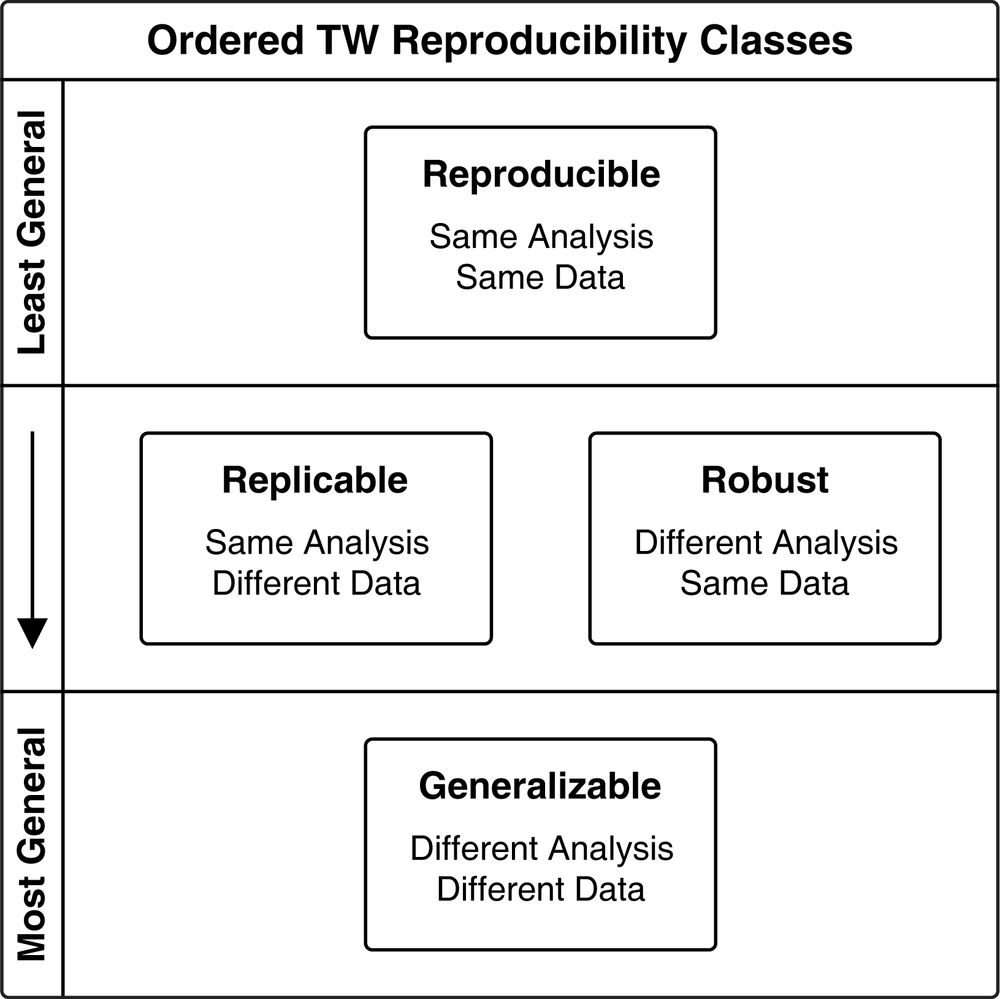
November 29, 2023 at 3:51 PM
QIIME 2 Provenance Replay auto-generates the code for creating QIIME 2 results from QIIME 2 results & builds "bioinformatics reproducibility supplements" for your papers. Our goal is to make it easy for you to perform reproducible, replicable & robust bioinformatics.
doi.org/10.1371/jour...
doi.org/10.1371/jour...
#QIIME2 2023.9 is now available! Exciting updates include an alpha release of our shotgun #metagenomics distribution, fully integrated provenance replay within the framework, batched decontam analysis, and more! Thank you contributors!!
forum.qiime2.org/t/qiime-2-20...
forum.qiime2.org/t/qiime-2-20...
October 20, 2023 at 6:49 PM
#QIIME2 2023.9 is now available! Exciting updates include an alpha release of our shotgun #metagenomics distribution, fully integrated provenance replay within the framework, batched decontam analysis, and more! Thank you contributors!!
forum.qiime2.org/t/qiime-2-20...
forum.qiime2.org/t/qiime-2-20...

