
📜: arxiv.org/abs/2412.17780
💻: huggingface.co/ChatterjeeLa...

📜: arxiv.org/abs/2412.17780
💻: huggingface.co/ChatterjeeLa...
@naturebiotech.bsky.social! 🌟 Submit your paper and join us in Singapore! 🇸🇬
Website: gembio.ai
Papers Due: February 3rd, 2025 📜
- I'm moving to Senior Applied Research Scientist in Digital Biology at NVIDIA (Jan '25)
- I'm starting a new lab at Duke as Visiting Assistant Prof (early '25)
Both roles focus on tackling hard problems in biological machine learning through collaborative research.
Long 🧵
FutureHouse's postdoctoral fellowship and indicate my lab as an option! 😃 $125k salary and access to all of their amazing resources! 🌟

FutureHouse's postdoctoral fellowship and indicate my lab as an option! 😃 $125k salary and access to all of their amazing resources! 🌟
www.forbes.com/sites/alexyo...

www.forbes.com/sites/alexyo...
#EndAxD Instagram Post: www.instagram.com/p/DC7sV2GPst...

#EndAxD Instagram Post: www.instagram.com/p/DC7sV2GPst...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Communications Biology! Here, we fine-tune the ESM-2 pLM to identify peptidic binding sites on target-interacting partner sequences. We fuse these "guide" peptides to E3 ubiquitin ligases to degrade disease-causing proteins! Take a read! :) www.nature.com/articles/s42...
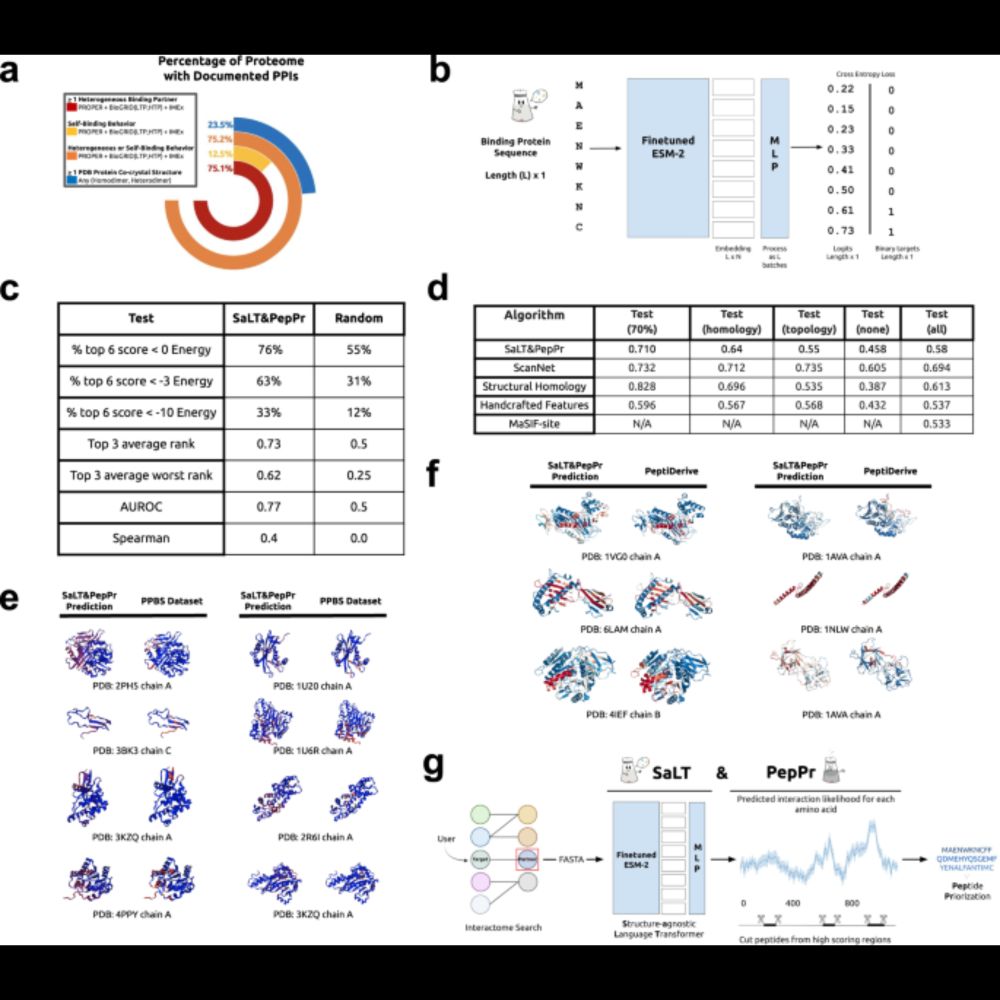
Communications Biology! Here, we fine-tune the ESM-2 pLM to identify peptidic binding sites on target-interacting partner sequences. We fuse these "guide" peptides to E3 ubiquitin ligases to degrade disease-causing proteins! Take a read! :) www.nature.com/articles/s42...

Fine-tunes ESM-2 network to achieve “target-conditioned de novo binder design from sequence alone”
arxiv.org/abs/2310.03842
huggingface.co/TianlaiChen/...

Fine-tunes ESM-2 network to achieve “target-conditioned de novo binder design from sequence alone”
arxiv.org/abs/2310.03842
huggingface.co/TianlaiChen/...

