Molecolab Pisa
@molecolabpisa.bsky.social
Computational Chemistry Research Group at the University of Pisa, Italy https://molecolab.dcci.unipi.it/
Carotenoid antennas 📡 for rhodopsins? Laura's new work reveals the structural origin of Carotenoid to Retinal energy transfer in the proton-pumping rhodopsin Kin4B8.
See the full news molecolab.dcci.unipi.it/eet-rhodopsi...
#openaccess doi.org/10.1039/D5SC...
See the full news molecolab.dcci.unipi.it/eet-rhodopsi...
#openaccess doi.org/10.1039/D5SC...
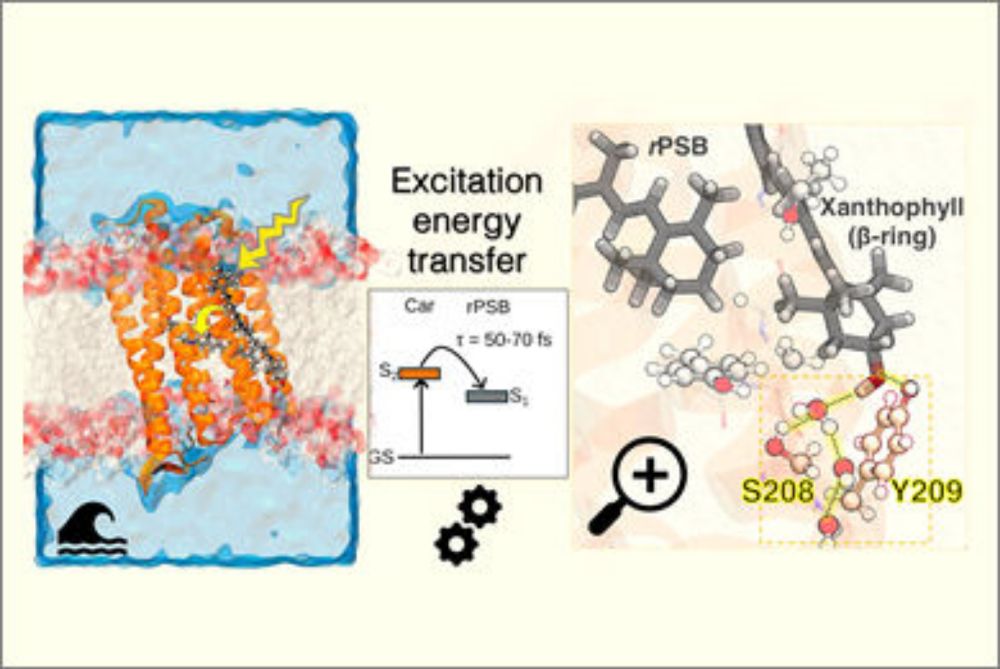
Structural and Spectroscopic Basis of Excitation Energy Transfer in Microbial Rhodopsins Binding Xanthophylls
Carotenoids are essential for light harvesting in certain microbial rhodopsins, with xanthorhodopsins that bind salinaxanthin being the most well-known example. But how do carotenoids that lack the ty...
molecolab.dcci.unipi.it
September 4, 2025 at 8:02 AM
Carotenoid antennas 📡 for rhodopsins? Laura's new work reveals the structural origin of Carotenoid to Retinal energy transfer in the proton-pumping rhodopsin Kin4B8.
See the full news molecolab.dcci.unipi.it/eet-rhodopsi...
#openaccess doi.org/10.1039/D5SC...
See the full news molecolab.dcci.unipi.it/eet-rhodopsi...
#openaccess doi.org/10.1039/D5SC...
Molecolab members at #ESPCongress2025 @photobiologyeurope.bsky.social !
molecolab.dcci.unipi.it/molecolab-es...
Lorenzo @lcupellini.bsky.social Laura and Piermarco
molecolab.dcci.unipi.it/molecolab-es...
Lorenzo @lcupellini.bsky.social Laura and Piermarco

MolecoLab at the 21st Congress of European Society for Photobiology, ESP-2025
MolecoLab was proudly represented at the 21st Congress of European Society for Photobiology last week in Bari (Italy), with three members contributing to key symposia on cutting-edge re...
molecolab.dcci.unipi.it
September 4, 2025 at 7:40 AM
Molecolab members at #ESPCongress2025 @photobiologyeurope.bsky.social !
molecolab.dcci.unipi.it/molecolab-es...
Lorenzo @lcupellini.bsky.social Laura and Piermarco
molecolab.dcci.unipi.it/molecolab-es...
Lorenzo @lcupellini.bsky.social Laura and Piermarco
New work is out!
We implemented linear response CASSCF in CFour, powered by the open-source Diaglib solver: we are now able to access absorption spectra & frequency-dependent properties on standard hardware.
doi.org/10.1021/acs....
#compchem #molecolab #casscf @tommasonottoli.bsky.social
We implemented linear response CASSCF in CFour, powered by the open-source Diaglib solver: we are now able to access absorption spectra & frequency-dependent properties on standard hardware.
doi.org/10.1021/acs....
#compchem #molecolab #casscf @tommasonottoli.bsky.social

An Efficient and Robust Implementation of CASSCF Linear Response Theory
We present a robust and efficient implementation of linear response theory for a Complete Active Space─Self-Consistent Field wave function. Our approach relies on the Cholesky Decomposition of the two-electron integrals, enabling the routine treatment of large molecular systems on standard hardware. It allows for the computation of both absorption energies and transition properties, as well as frequency-dependent molecular response functions. For both classes of properties, numerically stable and efficient algorithms have been developed. The capabilities of our implementation are demonstrated through the calculation of absorption spectra and molecular response properties of large systems with extended basis sets.
doi.org
August 29, 2025 at 4:16 PM
New work is out!
We implemented linear response CASSCF in CFour, powered by the open-source Diaglib solver: we are now able to access absorption spectra & frequency-dependent properties on standard hardware.
doi.org/10.1021/acs....
#compchem #molecolab #casscf @tommasonottoli.bsky.social
We implemented linear response CASSCF in CFour, powered by the open-source Diaglib solver: we are now able to access absorption spectra & frequency-dependent properties on standard hardware.
doi.org/10.1021/acs....
#compchem #molecolab #casscf @tommasonottoli.bsky.social
Mariastella's work on the electron transfer in the cryptochrome of birds has been selected as the front cover of today’s issue of
JPC Letters! @pubs.acs.org #MyACSCover doi.org/10.1021/acs.jpclett.5c01814 @lcupellini.bsky.social
JPC Letters! @pubs.acs.org #MyACSCover doi.org/10.1021/acs.jpclett.5c01814 @lcupellini.bsky.social

August 28, 2025 at 9:45 AM
Mariastella's work on the electron transfer in the cryptochrome of birds has been selected as the front cover of today’s issue of
JPC Letters! @pubs.acs.org #MyACSCover doi.org/10.1021/acs.jpclett.5c01814 @lcupellini.bsky.social
JPC Letters! @pubs.acs.org #MyACSCover doi.org/10.1021/acs.jpclett.5c01814 @lcupellini.bsky.social
Reposted by Molecolab Pisa
Last week I had the opportunity to present my poster at WATOC2025✨
It was a valuable occasion to share my work and engage with researchers in the field.
Many thanks to the organizers and participants for the stimulating event.
#WATOC2025 #computationalchemistry #machinelearning
It was a valuable occasion to share my work and engage with researchers in the field.
Many thanks to the organizers and participants for the stimulating event.
#WATOC2025 #computationalchemistry #machinelearning

June 30, 2025 at 9:12 AM
Last week I had the opportunity to present my poster at WATOC2025✨
It was a valuable occasion to share my work and engage with researchers in the field.
Many thanks to the organizers and participants for the stimulating event.
#WATOC2025 #computationalchemistry #machinelearning
It was a valuable occasion to share my work and engage with researchers in the field.
Many thanks to the organizers and participants for the stimulating event.
#WATOC2025 #computationalchemistry #machinelearning
Patrizia is back—and she brought cake! That's how you make a comeback. Thanks, @pamazzeo.bsky.social !

May 22, 2025 at 4:21 PM
Patrizia is back—and she brought cake! That's how you make a comeback. Thanks, @pamazzeo.bsky.social !
Patrizia's talk at the 2025 SIMPLAIX @simplaix.bsky.social workshop explaining ML/MM methods for solvent-dependent excited-state dymamics
#machinelearning #ML #compchem
#machinelearning #ML #compchem

May 7, 2025 at 4:16 PM
Patrizia's talk at the 2025 SIMPLAIX @simplaix.bsky.social workshop explaining ML/MM methods for solvent-dependent excited-state dymamics
#machinelearning #ML #compchem
#machinelearning #ML #compchem
Take a look at our new computational protocol presented in #JPCB to model excited-state lifetimes of Light-harvesting complexes!
doi.org/10.1021/acs....
Congrats to Chris for her first paper at Molecolab!
#compchem #theoreticalchemistry #photochemistry
doi.org/10.1021/acs....
Congrats to Chris for her first paper at Molecolab!
#compchem #theoreticalchemistry #photochemistry

A Computational Approach to Modeling Excitation Energy Transfer and Quenching in Light-Harvesting Complexes
Light-harvesting complexes (LHCs) play a critical role in modulating energy flux within photosynthetic organisms in response to fluctuating light. Under high light conditions, they activate quenching ...
doi.org
January 20, 2025 at 2:47 PM
Take a look at our new computational protocol presented in #JPCB to model excited-state lifetimes of Light-harvesting complexes!
doi.org/10.1021/acs....
Congrats to Chris for her first paper at Molecolab!
#compchem #theoreticalchemistry #photochemistry
doi.org/10.1021/acs....
Congrats to Chris for her first paper at Molecolab!
#compchem #theoreticalchemistry #photochemistry
Our computational study on a fatty acid photodecarboxylase is out in JACS Au #openaccess.
Check out what drives the electron transfer and decarboxylation in this exciting #photoenzyme!
pubs.acs.org/doi/10.1021/...
#QMMM #compchem #photobiocatalysis
Congrats to Giacomo and all authors!
Check out what drives the electron transfer and decarboxylation in this exciting #photoenzyme!
pubs.acs.org/doi/10.1021/...
#QMMM #compchem #photobiocatalysis
Congrats to Giacomo and all authors!

Protein-Driven Electron-Transfer Process in a Fatty Acid Photodecarboxylase
Naturally occurring photoenzymes are rare in nature, but among them, fatty acid photodecarboxylases derived from Chlorella variabilis (CvFAPs) have emerged as promising photobiocatalysts capable of performing the redox-neutral, light-induced decarboxylation of free fatty acids (FAs) into C1-shortened n-alka(e)nes. Using a hybrid QM/MM approach combined with a polarizable embedding scheme, we identify the structural changes of the active site and determine the energetic landscape of the forward electron transfer (fET) from the FA substrate to the excited flavin adenine dinucleotide. We obtain a charge-transfer diradical structure where a water molecule rearranges spontaneously to form a H-bond interaction with the excited flavin, while the FA’s carboxylate group twists and migrates away from it. Together, these structural modifications provide the driving force necessary for the fET to proceed in a downhill direction. Moreover, by examining the R451K mutant where the FA substrate is farther from the flavin core, we show that the marked reduction of the electronic coupling is counterbalanced by an increased driving force, resulting in a fET lifetime similar to the WT, thereby suggesting a resilience of the process to this mutation. Finally, through QM/MM molecular dynamic simulations, we reveal that, following fET, the decarboxylation of the FA radical occurs within tens of picoseconds, overcoming an energy barrier of ∼0.1 eV. Overall, by providing an atomistic characterization of the photoactivation of CvFAP, this work can be used for future protein engineering.
pubs.acs.org
December 20, 2024 at 11:10 AM
Our computational study on a fatty acid photodecarboxylase is out in JACS Au #openaccess.
Check out what drives the electron transfer and decarboxylation in this exciting #photoenzyme!
pubs.acs.org/doi/10.1021/...
#QMMM #compchem #photobiocatalysis
Congrats to Giacomo and all authors!
Check out what drives the electron transfer and decarboxylation in this exciting #photoenzyme!
pubs.acs.org/doi/10.1021/...
#QMMM #compchem #photobiocatalysis
Congrats to Giacomo and all authors!

