🧵(1/n)
www.cell.com/cell/fulltex...

careers.gene.com/us/en/job/20...

careers.gene.com/us/en/job/20...



@stanfordmedicine.bsky.social
is recruiting! We are looking for a postdoc at the interface of quantitative proteomics and G protein-coupled receptor (GPCR) biology: postdocs.stanford.edu/prospective/...
#TeamMassSpec #Proteomics #GPCR #Postdoc
@stanfordmedicine.bsky.social
is recruiting! We are looking for a postdoc at the interface of quantitative proteomics and G protein-coupled receptor (GPCR) biology: postdocs.stanford.edu/prospective/...
#TeamMassSpec #Proteomics #GPCR #Postdoc
#FunctionalGenomics #Seminar

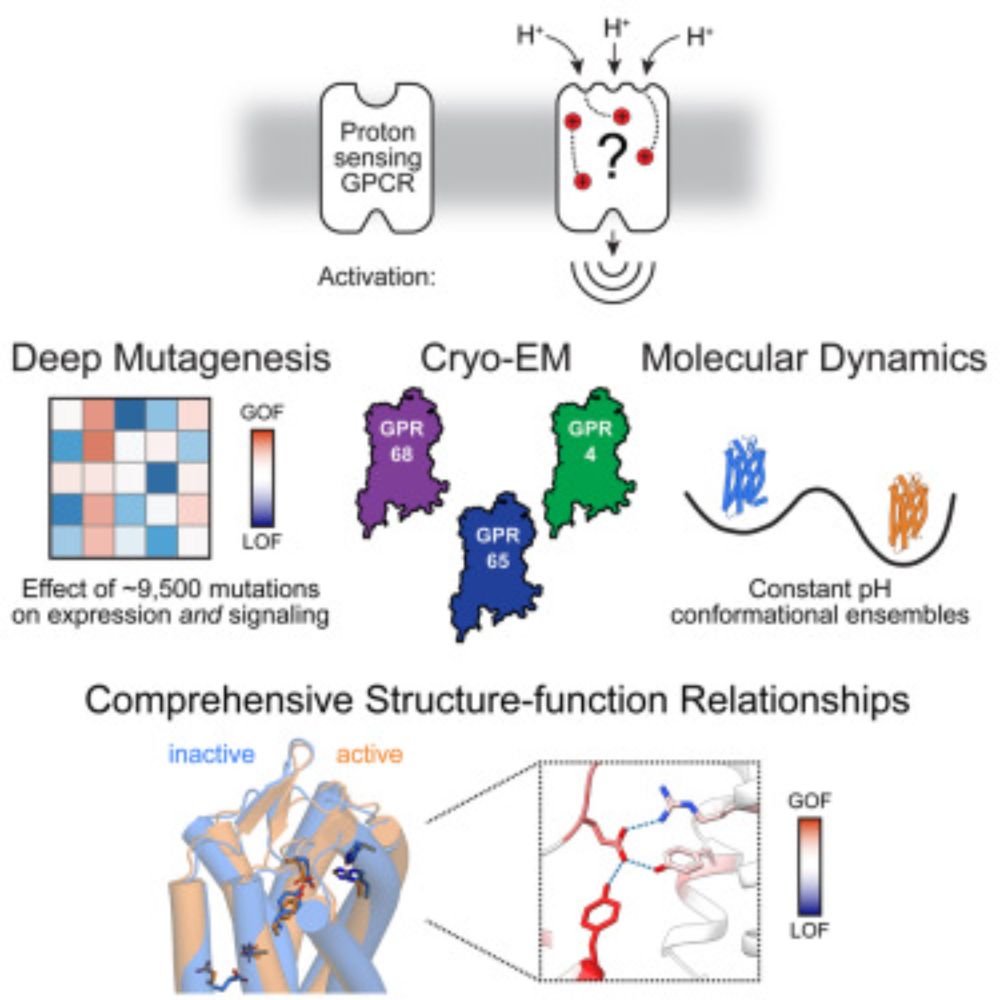
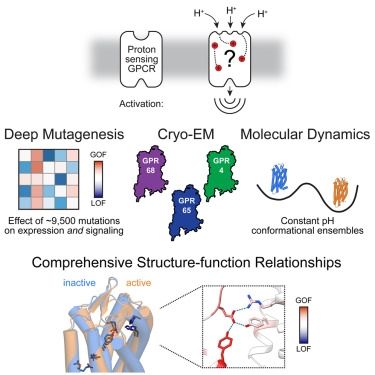
Mapping function to structure with deep mutational scanning to resolve the mechanisms of pH sensing GPCRs
www.sciencedirect.com/science/arti...
🧵(1/n)
www.cell.com/cell/fulltex...
🧵(1/n)
www.cell.com/cell/fulltex...
rdcu.be/d1OTf
rdcu.be/d1OTf

