Interest: Neurobiology, the evolution of the nervous system, GPCR deorphanisation, non-bilaterians.
rupress.org/jgp/article/...
rupress.org/jgp/article/...
www.nature.com/articles/s42...

www.nature.com/articles/s42...




www.nature.com/articles/s41...
This is a major output from our ERC-StG project Evocellmap @erc.europa.eu at @crg.eu
www.nature.com/articles/s41...
This is a major output from our ERC-StG project Evocellmap @erc.europa.eu at @crg.eu
with Yanez-Guerra et al.
https://www.biorxiv.org/content/10.1101/2025.04.18.649542v1
Trichoplax has tryptamine, tyramine, and phenethylamine receptors […]
[Original post on biologists.social]
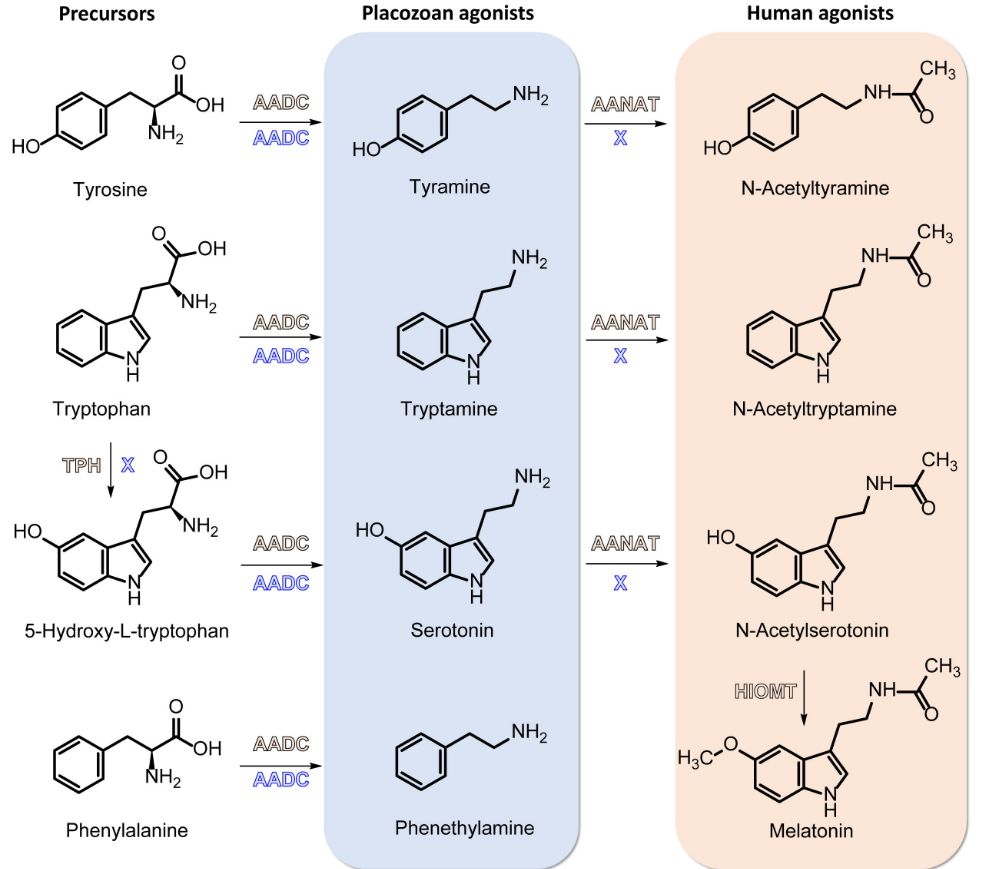
In the most recent work of my laboratory, we demonstrate that placozoans can respond to endogenous monoamines. Here you can look at the effects of tyramine, which increases the speed of placozoans. Tryptamine and Phenethylamine also have effects.
#Science #Placozoans #Monoamines

In the most recent work of my laboratory, we demonstrate that placozoans can respond to endogenous monoamines. Here you can look at the effects of tyramine, which increases the speed of placozoans. Tryptamine and Phenethylamine also have effects.
#Science #Placozoans #Monoamines
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
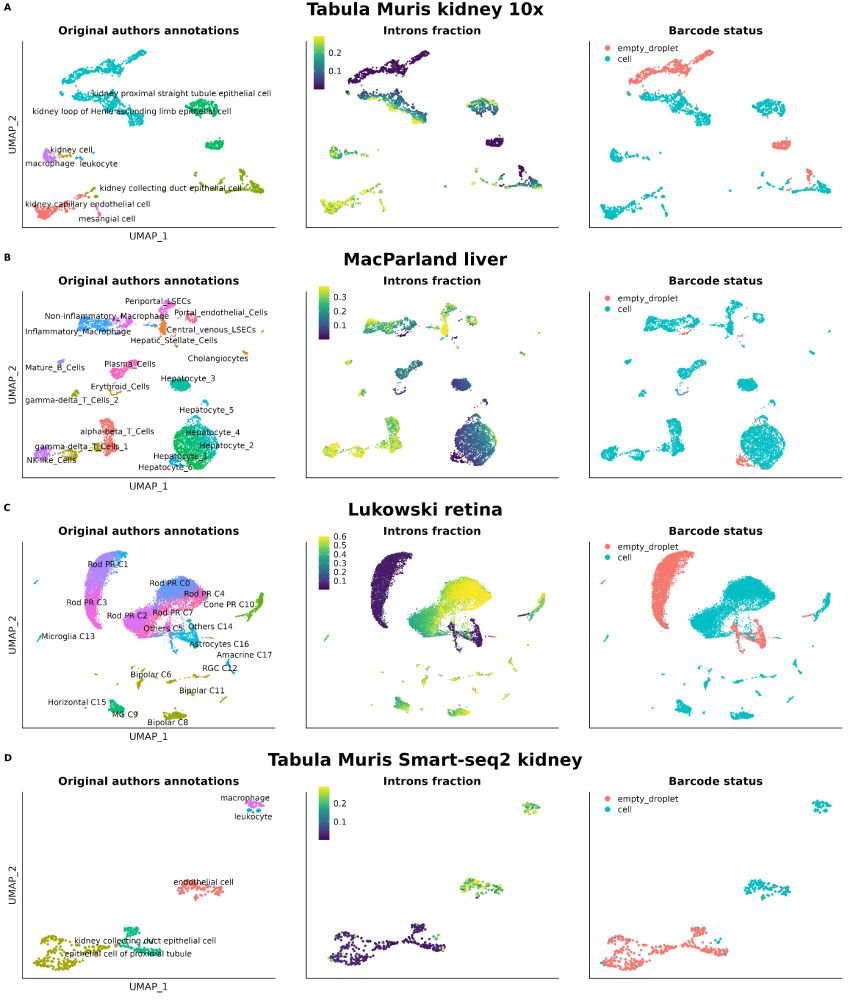
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8


