
From Belgium 🇧🇪 currently living in Cape Town 🇿🇦
#bioML #TeamMassSpec
Catch up on our blog and learn how we're helping to uncover new discoveries with improved peptide recall and more accurate error estimates. 🔍
#TeamMassSpec #Proteomics

Catch up on our blog and learn how we're helping to uncover new discoveries with improved peptide recall and more accurate error estimates. 🔍
#TeamMassSpec #Proteomics
Protein Language Model-Aligned Spectra Embeddings for De Novo Peptide Sequencing
https://www.biorxiv.org/content/10.1101/2025.10.01.679857v1
Protein Language Model-Aligned Spectra Embeddings for De Novo Peptide Sequencing
https://www.biorxiv.org/content/10.1101/2025.10.01.679857v1
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
#TeamMassSpec #Proteomics
arxiv.org/abs/2509.24952
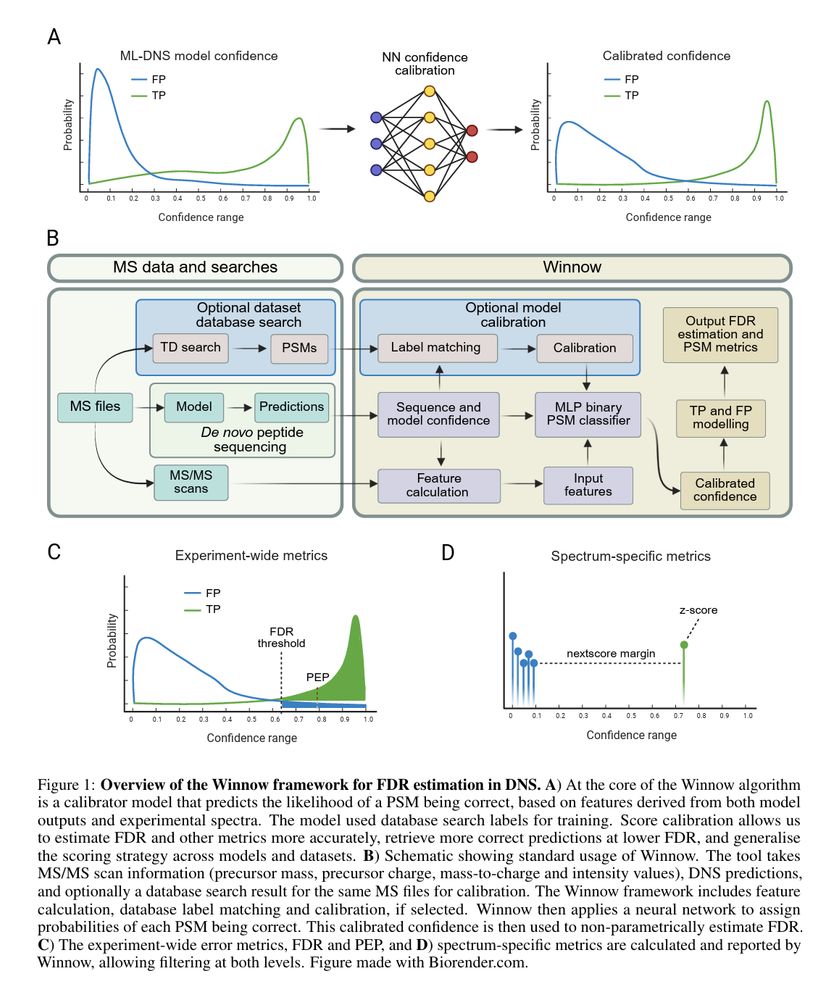
Improves de novo peptide sequencing by calibrating confidence scores with neural networks and estimating false discovery rates without decoy databases.




#TeamMassSpec #Proteomics
arxiv.org/abs/2509.24952
Improves de novo peptide sequencing by calibrating confidence scores with neural networks and estimating false discovery rates without decoy databases.




Improves de novo peptide sequencing by calibrating confidence scores with neural networks and estimating false discovery rates without decoy databases.
---
#proteomics #prot-preprint
---
#proteomics #prot-preprint
@instadeep.bsky.social: Winnow, our method for estimating false discovery rate in de novo peptide sequencing.

@instadeep.bsky.social: Winnow, our method for estimating false discovery rate in de novo peptide sequencing.
Check it out here: www.biorxiv.org/content/10.1...

Check it out here: www.biorxiv.org/content/10.1...

Check it out here: www.biorxiv.org/content/10.1...
✨ Machine Learning Foundations
✨ Generative Models & LLMs for African languages
All tutorial content will also be available online after the Indaba. Don’t miss out, subscribe here 👉 lnkd.in/eCgXRqsV




✨ Machine Learning Foundations
✨ Generative Models & LLMs for African languages
All tutorial content will also be available online after the Indaba. Don’t miss out, subscribe here 👉 lnkd.in/eCgXRqsV
A huge thank you to our expert presenters for sharing their knowledge: Luis Serrano, Dr. Arnu Pretorius and Yousra Farhan.




A huge thank you to our expert presenters for sharing their knowledge: Luis Serrano, Dr. Arnu Pretorius and Yousra Farhan.
The Women in Machine Learning (WIML) event was dedicated to an open discussion on the challenges women face in the field, and to exploring potential solutions.




The Women in Machine Learning (WIML) event was dedicated to an open discussion on the challenges women face in the field, and to exploring potential solutions.
De novo seq. IDs phos. sites. Transformer enhances analysis.
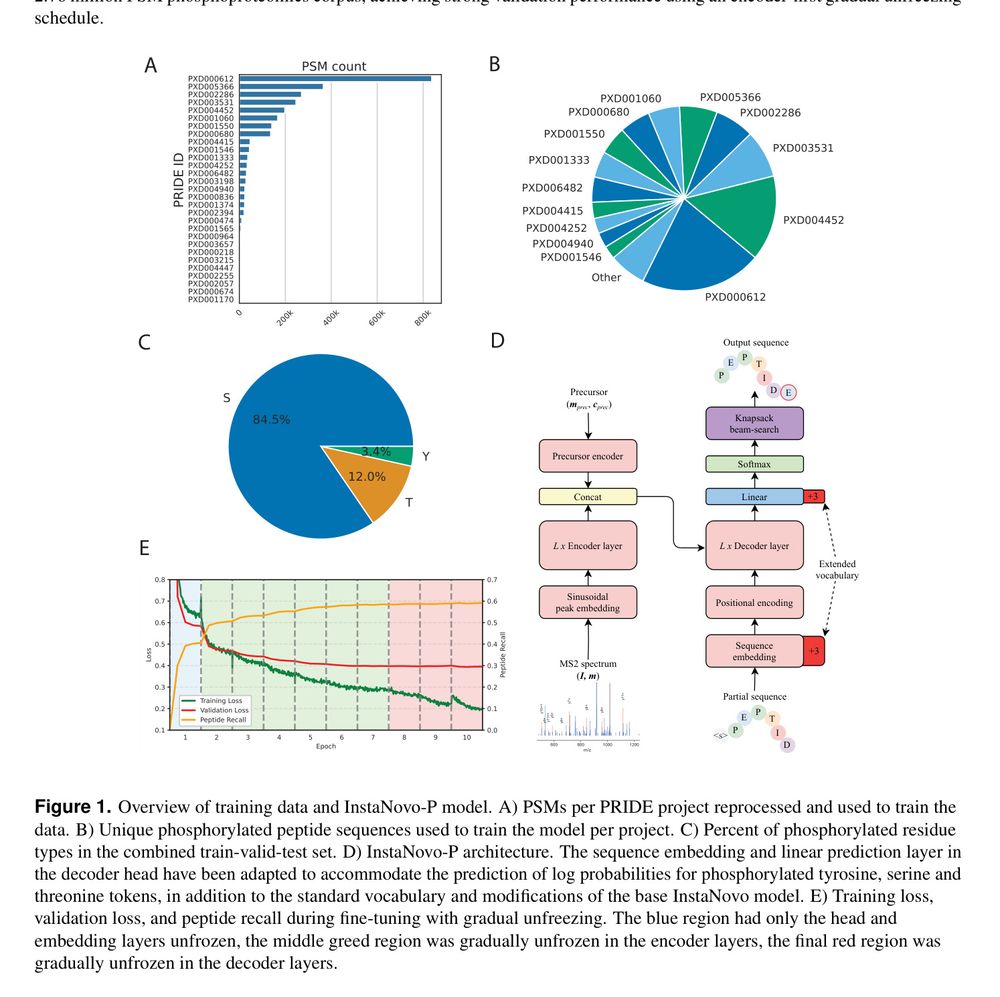



De novo seq. IDs phos. sites. Transformer enhances analysis.

---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
It’s fast, powerful, and shows how AI can help wet lab biology in very real ways! 🧪
Read and subscribe to get more like this plentyofroom.beehiiv.com/p/ai-protein...

It’s fast, powerful, and shows how AI can help wet lab biology in very real ways! 🧪
Read and subscribe to get more like this plentyofroom.beehiiv.com/p/ai-protein...
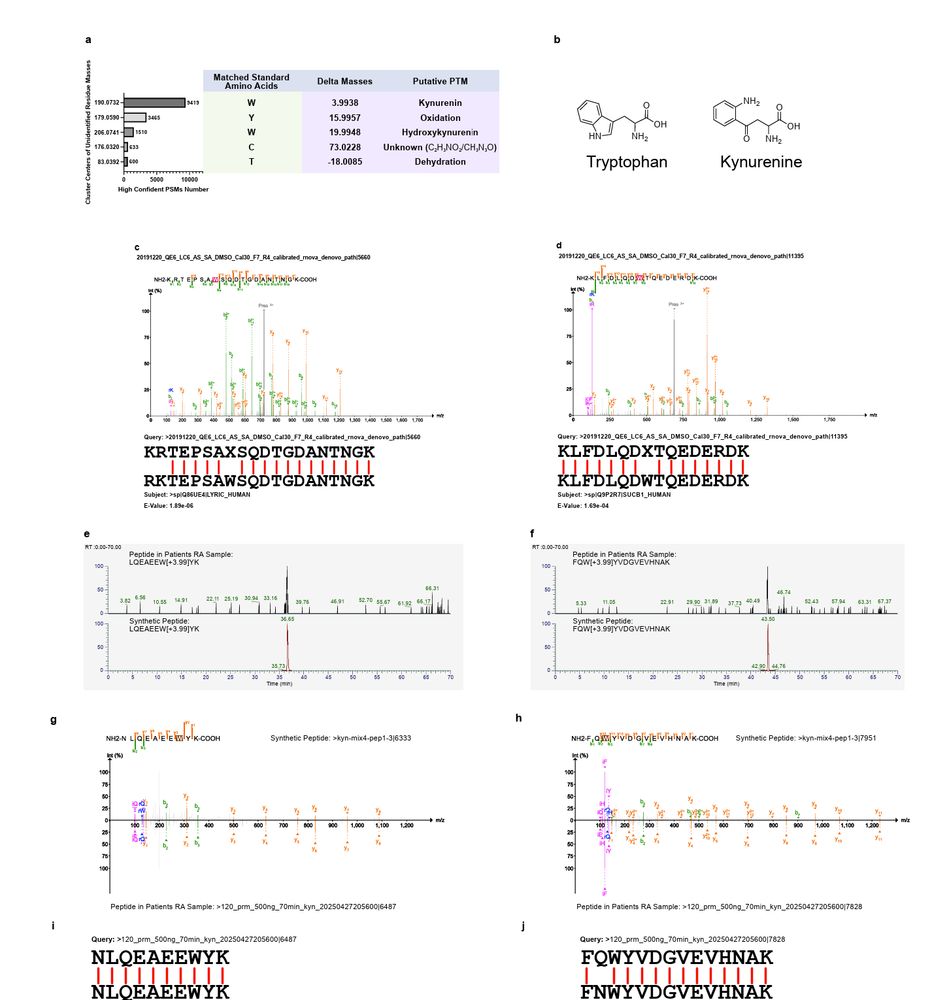
✅ 42% accuracy boost on HeLa proteome data
✅ 175% more peptides detected in immunopeptidomics (key for cancer and vaccine research)
✅ Reconstructs nanobodies and antibodies for therapeutic design
✅ Detects peptides from unknown microbes, venoms, and more🧪
✅ 42% accuracy boost on HeLa proteome data
✅ 175% more peptides detected in immunopeptidomics (key for cancer and vaccine research)
✅ Reconstructs nanobodies and antibodies for therapeutic design
✅ Detects peptides from unknown microbes, venoms, and more🧪
Here's how it works:
Here's how it works:
dx.doi.org/10.1101/2025...

dx.doi.org/10.1101/2025...
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
---
#proteomics #prot-preprint
---
#proteomics #prot-preprint





